FIG. 3. Binding of chimeric SRS-MTBD constructs to microtubules.
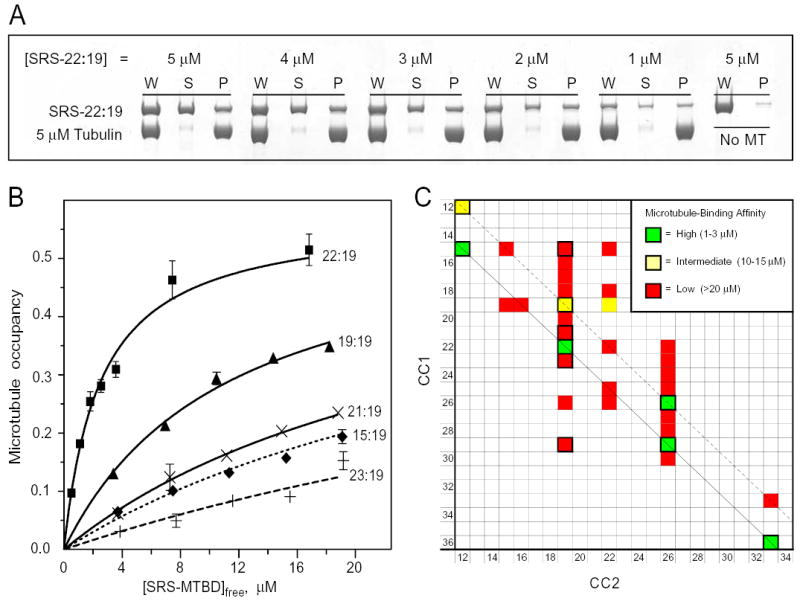
A, Polyacrylamide electrophoresis gels showing the binding of different concentrations of SRS-22:19 to microtubules. Binding was assayed by co-sedimentation after incubation with a suspension of 5 μM microtubules and the indicated concentrations of SRS-22:19 for 15 min at room temperature. See Methods for assay details. W, uncentrifuged sample, S, supernatant; P, pellet. A parallel sample of SRS-22:19 incubated and centrifuged with no microtubules was used as a blank (typically 1–2%). Recovery of SRS-MTBD and tubulin averaged (94 ± 10)%. B, Microtubule binding affinity of SRS-MTBD constructs with a fixed CC2 length of 19 amino acids and different lengths of CC1. Assays and gel electrophoresis were performed essentially as in Fig. 3A. Error bars indicate standard error of 2 replicate gels of the same microtubule-binding assay. Averages of affinity data from multiple independent preparations are given in text. C, Affinity of microtubule binding by SRS-MTBD constructs with different lengths of CC1 and CC2. The affinity of 33 constructs with CC1/CC2 lengths ranging from 12 to 36 amino acids was classified as high, medium or low by comparing the fraction of SRS-MTBD bound to microtubules in side-by-side assays with one or more of the constructs whose affinity had been assayed in detail as shown in Fig 3B. Assays were performed with 3 μM and 10 μM SRS-MTBD with 5 μM microtubules. The shortest constructs, SRS-12:12 and SRS-15:12, were less stable than the others and assays involving them have been corrected for presence of a non-binding aggregated form. Diagonal lines indicate constructs related to SRS-22:19 (solid black) and SRS-19:19 (dashed black). Cells with thick black border indicate constructs for which Rs has been determined (Supplemental data).
