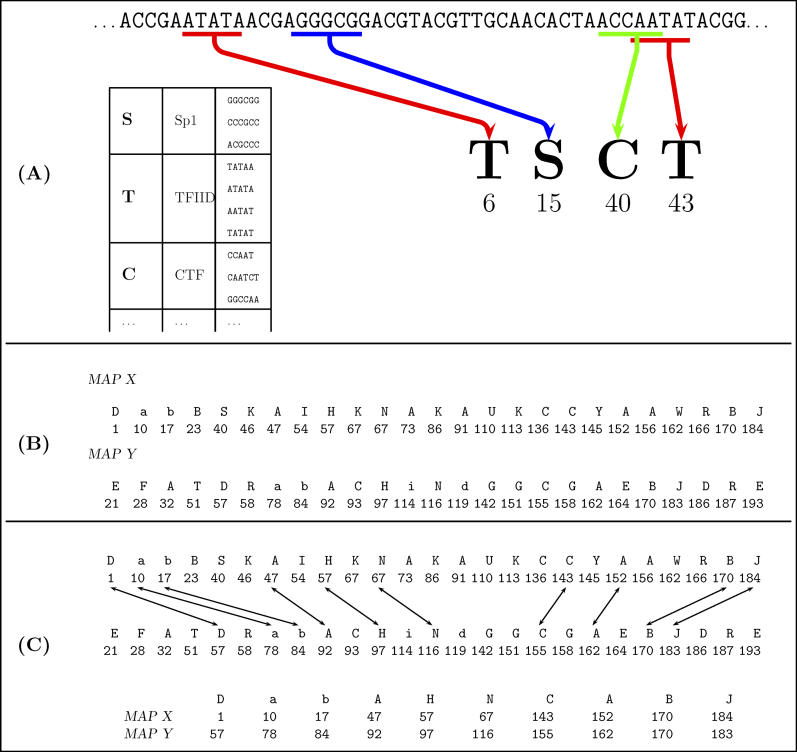
Figure 1. TF-Map Alignment of the Promoters of Two Hypothetical Co-Regulated Genes.
(A) The sequence of a promoter is searched for occurrences of known binding motifs for TFs. Matches are annotated with the position of the match in the primary sequence, and the label of the TF. Because TFs can bind to motifs showing no sequence conservation, labels of the same TF at different positions may correspond to different underlying nucleotide sequences. We refer here to these sequences of pairs (“label,” “position”) as TF-maps. TF-maps are actually more complicated. First, we do not only register the position of each match, but also its length. Second, while in the example here, sequence motifs are associated to TFs by means of a (binary) look-up table, in our work we have instead used collections of PWMs. Matches to TFBSs are thus scored, and this score is also registered.
(B) TF-map of the promoter region of two hypothetically co-regulated genes X and Y. Each letter corresponds to a different TF. We assume that 200 nucleotides upstream of the annotated TSS have been considered, with position 1 corresponding to position −200 from the TSS.
(C) Global pairwise alignment of the two co-regulated genes X and Y. Only positions with identical labels can be aligned. Essentially, the alignment finds the longest common substring constrained to maximizing the sum of the scores (unpublished data) of the aligned positions, and minimizing the differences in the distances on the primary sequence between adjacent aligned positions.