Abstract
tRNase Z, which can endonucleolytically remove pre-tRNA 3′-end trailers, possesses the signature His domain (HxHxDH; Motif II) of the β-lactamase family of metal-dependent hydrolases. Motif II combines with Motifs III–V on its carboxy side to coordinate two divalent metal ions, constituting the catalytic core. The PxKxRN loop and Motif I on the amino side of Motif II have been suggested to modulate tRNase Z activity, including the anti-determinant effect of CCA in mature tRNA. Ala walks through these two homology blocks reveal residues in which the substitutions unexpectedly reduce catalytic efficiency. While substitutions in Motif II can drastically affect kcat without affecting kM, five- to 15-fold increases in kM are observed with substitutions in several conserved residues in the PxKxRN loop and Motif I. These increases in kM suggest a model for substrate binding. Expressed tRNase Z processes mature tRNA with CCA at the 3′ end ∼80 times less efficiently than a pre-tRNA possessing natural sequence of the 3′-end trailer, due to reduced kcat with no effect on kM, showing the CCA anti-determinant to be a characteristic of this enzyme.
Keywords: pre-tRNA, 3′ processing endonuclease, tRNase Z, homology blocks
INTRODUCTION
Transfer RNAs are central to protein synthesis. All tRNAs are transcribed as precursors and processed by removal of extra sequences from their ends before they can be aminoacylated and used in translation. The 5′-end leader is removed by a widely conserved enzyme, RNase P (for review, see Xiao et al. 2002). All mature tRNAs have CCA at the 3′ end, and there are two different paths to this end (for review, see Mörl and Marchfelder 2001). In Escherichia coli, CCA is transcriptionally encoded, and the 3′-end trailer is first cut by the endonuclease RNase E and then trimmed by a chorus of exonucleases (Li and Deutscher 1996, 2002; Ow and Kushner 2002). In this organism, the CCA-adding enzyme is nonessential; the slow growth phenotype of the knockout suggests that it is important for repair of damaged tRNA 3′ ends (Zhu and Deutscher 1987). In eukaryotes, archaea, and many bacteria in which CCA is not transcriptionally encoded—on the other hand—the CCA-adding enzyme is essential (Aebi et al. 1990). In these cases, in a widely conserved, efficient, and precise mechanism for pre-tRNA 3′ end maturation, a pre-tRNA 3′ end processing endonuclease, tRNase Z, cleaves the 3′-end trailer directly following the discriminator (the last unpaired nucleotide following the 7-bp acceptor stem), leaving a 3′ OH to which CCA can be directly added (Castaño et al. 1985; Frendewey et al. 1985; Chen and Martin 1988; Marchfelder et al. 1990; Nashimoto 1992; Levinger et al. 1995; for reviews, see Levinger et al. 2004b; Vogel et al. 2005). Interestingly, tRNA precursors with and without transcriptionally embedded CCA coexist in Bacillus subtilis, and the two pathways for pre-tRNA 3′ end maturation are used selectively on the appropriate substrates (Pellegrini et al. 2003). In this and other cases, CCA of mature tRNA is an anti-determinant that prevents mature tRNA from recycling through tRNase Z (Nashimoto 1997; Mohan et al. 1999). This has not been universally observed, however (Schiffer et al. 2003; see Discussion). In Thermotoga maritima, a bacterial species in which all but one of the pre-tRNAs have transcriptionally encoded CCA and important pre-tRNA 3′-end exonucleases are absent, tRNase Z cleaves atypically after CCA instead of after the discriminator (Minagawa et al. 2004).
tRNase Z is present in a short form (tRNase ZS) in bacteria and archaea and in a long form (tRNase ZL) in eukaryotes (Schiffer et al. 2002). tRNase ZL apparently evolved as a gene duplication of tRNase ZS in which the active site was retained in the carboxy half, leaving the amino half of tRNase ZL free to diverge (Tavtigian et al. 2001). For example, a flexible domain looped out from the globular core of tRNase ZS (de la Sierra-Gallay et al. 2005) that is involved in pre-tRNA binding (Schilling et al. 2005) was retained in the amino half and lost from the carboxy half of tRNase ZL. Some eukaryotes (Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster) have only tRNase ZL, which, in the case of the fruit fly, is involved in both nuclear and mitochondrial pre-tRNA maturation (Dubrovsky et al. 2004), while some (e.g., Arabidopsis thaliana, Homo sapiens, and other mammals) have both tRNase ZL and tRNase ZS.
Conserved homology blocks in the carboxy half of tRNase ZL align with blocks in tRNase ZS. Motif II consists of the His domain (HxHxDH), the signature sequence of the β-lactamase family of metal-dependent hydrolases, which, in collaboration with two other histidines and another aspartate (Motifs III–V), coordinates two divalent metals, constituting the active site. Two additional homology blocks on the amino side of the His domain (the PxKxRN loop and Motif I) have been suggested to modulate tRNase Z activity (Minagawa et al. 2004; de la Sierra-Gallay et al. 2005). Two residues in the Motif I region of T. maritima tRNase Z were suggested to be involved in its unusual selection of cleavage site and, by inference, the CCA anti-determinant (Minagawa et al. 2004). Alternatively, the PxKxRN loop could be involved in CCA anti-determination in B. subtilis tRNase Z (de la Sierra-Gallay et al. 2005); this loop is absent from T. maritima tRNase Z due to truncation of its amino end. To evaluate these hypotheses concerning regions and residues potentially involved in modulating tRNase Z activity, we analyzed the effects of Ala substitutions through the PxKxRN loop and Motif I of D. melanogaster tRNase Z on pre-tRNAHis 3′ end processing. Additionally, to quantitate the CCA anti-determinant effect, substrates possessing natural sequence of pre-tRNA and CCA following the discriminator were analyzed in parallel using expressed, affinity-purified tRNase Z.
RESULTS
Homology blocks on the amino side of the His domain (Motif II)
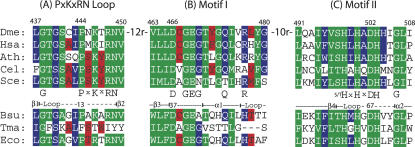
Multiple sequence alignments of tRNase ZL and tRNase ZS highlight the relationship of tRNase Z with other members of the β-lactamase family of metal-dependent hydrolases (Tavtigian et al. 2001; Schiffer et al. 2002; Takaku et al. 2003; Dubrovsky et al. 2004; Schilling et al. 2005) and the homology blocks most directly involved in catalysis (Motifs II–V in the terminology of Ishii et al. 2005). The PxKxRN loop and Motif I, two homology blocks on the amino side of Motif II (but still within the carboxy half of tRNase ZL) (Fig. 1A,B), are investigated herein, and the recently analyzed His domain (Motif II) (Fig. 1C; Zareen et al. 2005) is included to help establish context. Five eukaryotic tRNase ZLs (above) and three bacterial tRNase ZSs (below) are shown in the alignments. Numbers above the top panels refer to the residues in fruit fly tRNase Z (a long form) (see Materials and Methods; Zareen et al. 2005) in which the central histidine of Motif II is H500 (Fig. 1C). Residue numbers and secondary structure elements directly above the tRNase ZS alignments refer to B. subtilis tRNase Z. The most conserved residues are in green, semiconserved residues are in red, and similar residues (e.g., large hydrophobics) are in blue. These homology blocks are observed in all species, except that T. maritima tRNase Z has no PxKxRN loop (Fig. 1A), as noted before (de la Sierra-Gallay et al. 2005).
FIGURE 1.
Alignment of selected homology blocks in tRNase ZL and tRNase ZS proteins. (A) The PxKxRN loop, (B) the region including Motif I, and (C) the region surrounding Motif II. The top five aligned sequences are tRNase ZL’s from Drosophila melanogaster, Homo sapiens, Arabidopsis thaliana, Caenorhabditis elegans, and Saccharomyces cerevisiae (accession nos. Q8MKW7, NP_060597.3, AAM51378, O4476, and NP013005.1, respectively). The bottom three sequences are tRNase ZS’s from B. subtilis, T. maritime, and E. coli (accession nos. P54548, NP_228673, and ZP_00726790, respectively). Designations below the tRNase ZL panels indicate the consensus, and coloring of residues indicates the extent of homology (green, identical; red, conserved in most cases; purple, similar). Numbering of selected residues above tRNase ZL panels is for fruit fly tRNase ZL based on the presumed translation initiation at an internal methionine (r24) as previously described in Zareen et al. 2005. Spacing between the three homology blocks presented is also conserved in the five tRNase ZL’s. Residue numbers and designations above the bottom panel (indicating the positions of the secondary structure elements within the homology blocks) are based on Bsu tRNase ZS (de la Sierra Gallay et al. 2005).
Correlation of homology blocks with the B. subtilis tRNase Z structure model allows useful comparisons to be made between tRNase ZL and tRNase ZS
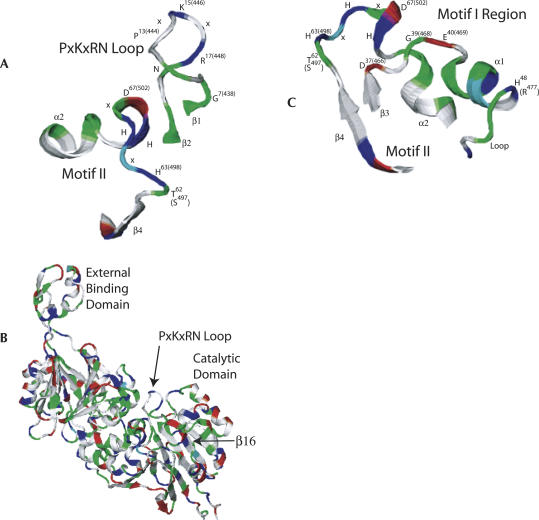
The structures of bacterial tRNase Zs (from B. subtilis, T. maritima, and E. coli) (de la Sierra-Gallay et al. 2005; Ishii et al. 2005; Kostelecky et al. 2006) show the same fold, two opposing clusters of the secondary structure elements ββββαβαβαβ in each subunit, in which the first β-strand is contributed by β16. Composite views adapted from the model of B. subtilis tRNase Z (Fig. 2A–C, displayed using VMD from PDB ID 1Y44; de la Sierra-Gallay et al. 2005) represent the contribution that could be made by the PxKxRN loop and Motif I (potential regulatory domains) and Motif II to tRNase Z catalysis.
FIGURE 2.
Views of tRNase Z taken from the B. subtilis structure (de la Sierra-Gallay et al. 2005; PDB ID 1Y44). Motif II and the other active site residues (Motifs III–V) are buried in the catalytic domain of the A subunit, and the PxKxRN loop appears to cover the entrance to the active site. (A) PxKxRN loop and Motif II. (B) entire Bsu. tRNase Z structure model in the same front view as in A with the external binding domain in the upper left. (C) Motif I region and Motif II; the best view was obtained by rotating ∼180° so that the external binding domain is in the upper right (a back view). The amino acid, α-helix, and β-sheet nomenclature refer to the B. subtilis structure. Residue numbers in parentheses refer to the fruit fly protein.
The alignments (Fig. 1) suggest that fundamental structural aspects (β-sheet, α-helix, loop size, and geometry) of the three blocks presented are conserved between their locations in the carboxy half of tRNase ZL and the catalytic (A) subunit of B. subtilis tRNase ZS. The lack of structural information from the amino half of tRNase ZL prevents use of the dimer interface of tRNase ZS to develop a model for catalysis by tRNase ZL, however, limiting detailed application of the tRNase ZS model to one active site (Motif II–V) in collaboration with one PxKxRN loop and one Motif I region.
From a variety of viewing angles the PxKxRN (β1–β2) loop appears to partially cover a cavernous opening to the active site (Motifs II–V) like a flap (Fig. 2B; de la Sierra-Gallay et al. 2005). Characteristics attributed to this loop include rigidity conferred by P13, a conserved surface lysine (K15), and the ability to discriminate against C74 of mature tRNA as the first nucleotide of a CCA anti-determinant (de la Sierra-Gallay et al. 2005). Motif I is a single aspartate (D466 in D. melanogaster tRNase ZL) at the start of the β3–β1 loop (D37 in B. subtilis tRNase Z [de la Sierra-Gallay et al. 2005]; and D25 in T. maritima tRNase Z [Ishii et al. 2005]), which is conserved in both L and S tRNase Zs (Fig. 1B). D37, found at the periphery of important residues surrounding the inner circle of Motifs II–V, which directly bind the two divalent metals (de la Sierra-Gallay et al. 2005), was essential for tRNase Z activity (Minagawa et al. 2004). The region including Motif I consists of β-strand, loop, α-helix, and loop (Figs. 1B, 2C). Two residues in α1 were suggested to determine the unusual cleavage site of T. maritima tRNase Z (after transcriptionally embedded CCA) (Minagawa et al. 2004), corresponding in position to D. melanogaster Y472 and Q474 (based on alignments in Fig. 1B). We therefore performed a detailed mutational analysis of the PxKxRN loop and Motif I region.
Expressed affinity purified soluble tRNase Z
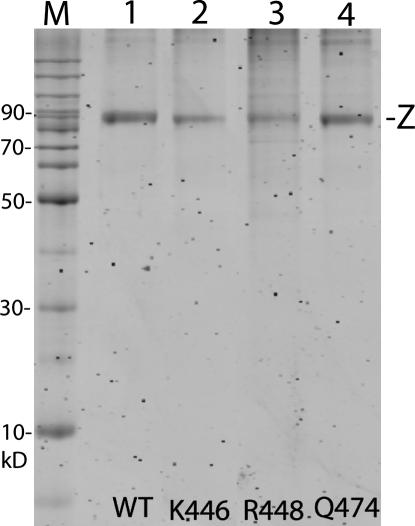
In both Figure 1 and Figure 2, A and C, the residues displayed are those that were subjected to Ala-scanning analysis (14 in the PxKxRN loop and 18 in the Motif I region). Wild-type and variant fruit fly tRNase Z's were constructed, baculovirus-expressed, and affinity-purified (Fig. 3; Zareen et al. 2005; see Materials and Methods). Yield and purity of soluble tRNase Z were sufficient to perform the efficiency and kinetic experiments. In Figure 3, ∼0.5 μg of each protein was loaded from the first step of a dilution series. The values for enzyme concentration used to calculate processing efficiency and kcat were refined with protein concentrations determined by scanning these protein gel lanes (Zareen et al. 2005; see Materials and Methods). The substitutions illustrated and several others in the PxKxRN loop and Motif I have substantial effects on processing efficiency and kinetics (see below).
FIGURE 3.
Baculovirus expression of fruit fly tRNase Z. Wild-type and mutant fruit fly tRNase Zs (indicated with –Z to the right of gel panel) were affinity-purified, electrophoresed on a 10% polyacrylamide SDS gel, stained with Sypro orange (Molecular Probes), and scanned for fluorescence with a Typhoon imager. The variant tRNase Z's presented (K446A, R448A, and Q474A, as designated below panel) were selected based on their effects on processing (see Fig. 3). Approximately 0.5 μg of each tRNase Z sample was loaded. The enzyme concentrations used in kinetic experiments were determined from the protein gel lanes with ImageQuant to obtain kcat from Vmax. Size designations are given for the marker lane to the left of the panel. Fruit fly tRNase Z displays an apparent molecular weight of ∼90 kDa, slightly larger than its predicted molecular weight of 83 kDa.
Kinetics of pre-tRNAHis processing
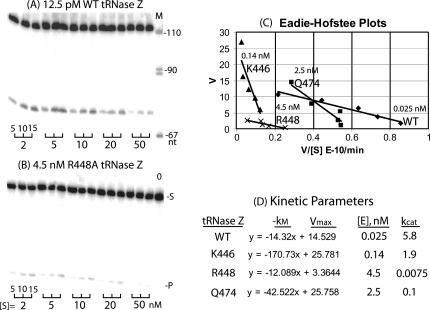
Methods and data quality of processing kinetic analysis are illustrated in Figure 4. Fruit fly pre-tRNAHis, the tRNase Z substrate used throughout (Frendewey et al. 1985; Levinger et al. 1995), is a 108-nt transcript consisting of a 72-nt tRNA (to the discriminator) and a 36-nt 3′-end trailer (substrate and tRNA product bands are marked on the right side of Fig. 4B). Processing efficiencies of enzymes with single residue substitutions were evaluated using a range of enzyme concentrations with labeled substrate and no added unlabeled substrate (data not shown; see Materials and Methods; Zareen et al. 2005). Concentrations of tRNase Zs with impairing substitutions were thereby adjusted to approximately match the overall extent of processing in the kinetic analysis. For example, tRNase Z with the R448A substitution was used at a concentration of 4.5 nM, ∼300 times more concentrated than the 12.5 pM wild-type tRNase Z (Fig. 4, cf. B and A). Increasing variant enzyme concentration could have unknown effects in addition to accelerating the assay, but the reactions catalyzed by enzyme variants with impairing substitutions all clearly fit the Michaelian model (Fig. 4C; data not shown).
FIGURE 4.
Kinetics of processing with wild-type and variant tRNase Z. Mutations analyzed are the same as in Figure 2. (A,B) The processing in a kinetic experiment with wild-type and R448A tRNase Z. Reactions were sampled after 5, 10, and 15 min as designated by the brackets below the data panels. The concentrations of tRNase Z (12.5 pM for wild type and 4.5 nM for R448A, designated above the panels) were chosen to produce a similar extent of processing (% product/minute of reaction, equivalent to V/[S]). The substrate concentrations, adjusted by adding unlabeled fruit fly pre-tRNAHis, were 2, 5, 10, 20, and 50 nM, as indicated below the panels. (C) Eadie-Hofstee plots for wild type, K446, R448, and Q474. Concentrations of enzymes used are indicated on the plot. (D) Kinetic parameters from the data presented in A–C. Equations presented for the enzymes (columns labeled –kM, Vmax, with units of × 10−9 M and × 10−11 M/min., respectively) are taken from the Excel plots (C). kcat is calculated from Vmax using the value for [E] determined from the protein gel lane for each mutant (Fig. 1) based on the dilution factor used in the experiment.
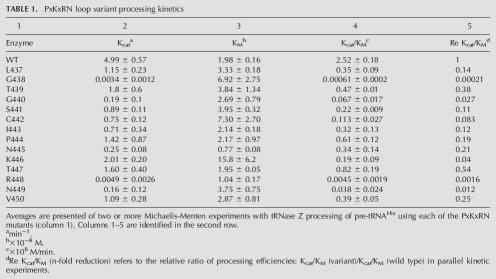
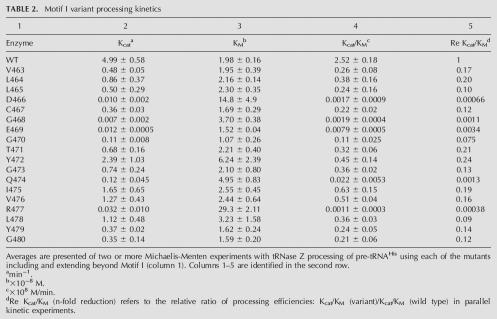
Eadie-Hofstee plots for four variants are superimposed in Figure 4C. Interestingly, two of the variants presented here (K446A in the PxKxRN loop and Q474A in Motif I) display a kM significantly higher than that of the wild-type enzyme (slope in Fig. 4C), which was not observed with any of the substitutions in Motif II (Zareen et al. 2005). Vmax (the intercepts on the ordinate in Fig. 4C) and the concentration of enzyme used in each reaction (shown in Fig. 4A–C and in the fourth column of D) were used to calculate kcat (rightmost column in Fig. 4D). In these representative examples of variant processing kinetics, in addition to the effects on kM, variant kcats were all reduced (three-, ∼60-, and 800-fold in the case of K446A, Q474A, and R448A, respectively). These results (and additional experiments performed at least twice for each variant enzyme; data not shown) were used to determine mean kcat, kM, and catalytic efficiency (kcat/kM) including the standard errors for all measurements, and the effect on catalytic efficiency relative to wild type (Tables 1, 2, columns 2–5).
TABLE 1.
PxKxRN loop variant processing kinetics
TABLE 2.
Motif I variant processing kinetics
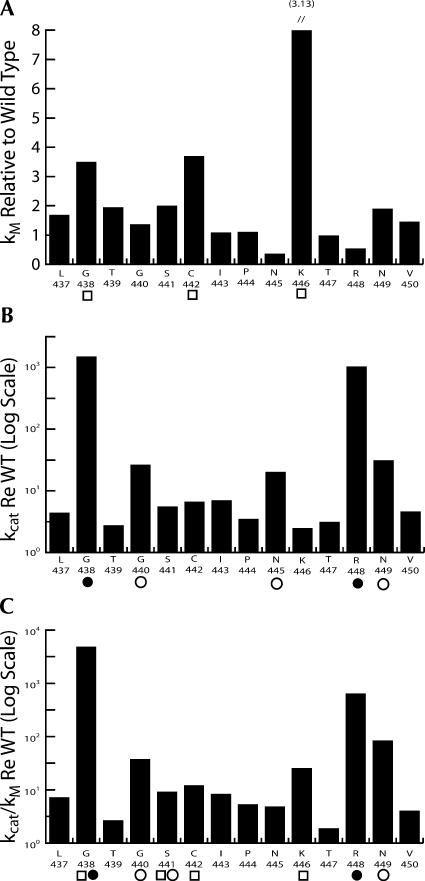
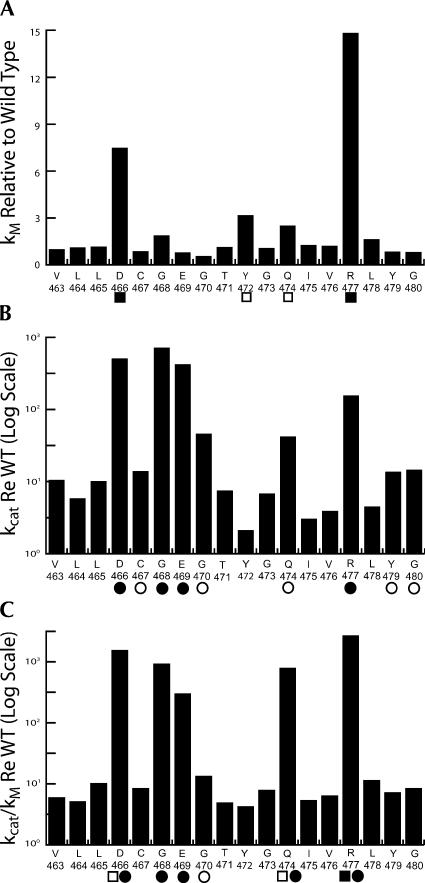
Results are presented graphically in Figures 5 and 6. In the bar graphs, kM relative to wild type (ordinate, linear scale) increased by up to eightfold for substitutions in the PxKxRN loop (K446A) (abscissa in Fig. 5A; cf. Fig. 4C,D and Table 1) and by up to 15-fold for substitutions in Motif I (R477A) (see Table 2; Fig. 6A). The relative effects of the substitutions on kcat and kcat/kM are presented using an inverse log scale (Figs. 5, 6, ordinate in panels B,C) to display the changes that reduce processing efficiency (relative increase in kM and decrease in kcat) in the same (positive) direction and because the effect of a single substitution on kcat can be up to 1000-fold greater than its effect on kM (e.g., G438A and R448A in the PxKxRN loop) (Fig. 5). In the most extreme cases (G438A in the PxKxRN loop [Table 1; Fig. 5C]; D466A and R477A in the Motif I region [Table 2; Fig. 6C]) processing efficiency was reduced between 1000- and 10,000-fold, comparable to the greatest reductions found with substitutions in Motif II (Zareen et al. 2005). Several mutants have been observed in which Ala substitution impairs reaction efficiency between 1000- and 10,000-fold, including D466, H500, and D502 (Zareen et al. 2005; this report), which were found by others to be essential for activity (Minagawa et al. 2004; Spath et al. 2005). These discrepancies could arise from differences in expression systems, assay methods, reaction optimization, and the form of tRNase Z (L or S). Expression and assay methods reported here allow the kinetic parameters to be obtained from mutant tRNAse Zs impaired by up to 105-fold relative to wild type. Some of the substitutions have slight effects, but both kcat and kcat/kM were reduced relative to wild type in all cases (the values on the ordinate in panels B,C of Figs. 5, 6 are all positive).
FIGURE 5.
Graphical representation of the effects of substitutions in the PxKxRN loop on tRNase Z processing. (A) kM relative to wild type (from column 3 in Table 1) is presented on a linear scale. The wild-type residues substituted with alanine in the PxKxRN loop are identified on the abscissa. The line and the bar above each value on the bar graph indicate the standard error (from Table 1). (B) The inverse of kcat relative to wild type (n-fold decrease in kcat, taken from column 2 in Table 1) is presented on a log scale. (C) The inverse of kcat/kM relative to wild type (taken from column 5 in Table 1) is presented on a log scale. Conserved residues (taken from Fig. 1A) that affect one or more of the kinetic parameters are presented with symbols (□,○ for the intermediate effects on kM and kcat, respectively; X,@ for the greatest effects). Error bars are not presented in B and C but can be obtained from Table 1.
FIGURE 6.
Graphical representation of the effects of substitutions in the Motif I region. The presentation of kM, kcat, and reaction efficiency (kcat/kM) relative to wild type in A–C and use of the symbols □,○,■,● is the same as in Figure 5.
Unlike the Motif II substitutions that reduce catalytic efficiency without affecting kM, substitutions in the PxKxRN loop and Motif I region can affect kcat, kM or both, as indicated by the symbols below the abscissas in Figures 5 and 6 (□,▪: moderate or great increase in kM, respectively; ○,•: moderate or great decrease in kcat), as follows: G438A and C442A cause a moderate (three- to fourfold) increase in kM and K446A causes a larger (about eightfold) increase in kM (PxKxRN loop) (Table 1; Fig. 5A); D466A and R477A (Motif I region) (Table 2; Fig. 6A) also cause large increases in kM (7.5- and 15-fold, respectively). The moderate and large decreases in kcat in the PxKxRN loop arise from the substitutions G440A, N445A, and N449A (between 100- and 1000-fold) and G438A and R448A (>1000×), respectively (Table 1; Fig. 5B), and those in the Motif I region arise from C467A, G470A, Q474A, Y479A, and G480A (100- to 1000-fold) and D466A, G468A, E469A, and R477A (>1000-fold) (Table 2 and Fig. 6B, respectively).
In the carboxy half of the PxKxRN loop, the order of importance of the conserved residues is P444 < K446 < R448 > N449 (Fig. 5). The effect of the R448A substitution on catalytic efficiency is 10 times greater than that of K446A; K446A acts principally through an increase in kM while R448A acts only by causing a decrease in kcat. Interestingly, substitution of R448 causes a reproducible decrease in kM in addition to the large decrease in kcat. Substitution of N445 causes an even larger decrease in kM; these are the only two residues that do so.
CCA at the 3′ end of mature tRNA is a tRNase Z anti-determinant that reduces kcat 80-fold
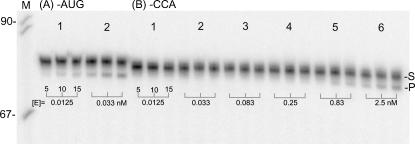
The efficiencies with which mature tRNA with CCA at the 3′ end and pre-tRNA with the short sequence –AUG (taken from sequence of the natural pre-tRNAHis 3′-end trailer) can be processed by tRNase Z were compared. The ∼1-nt greater length of the –AUG substrate (Fig. 7; see Materials and Methods) should not affect the results or interpretation of the –CCA anti-determinant experiments; the first three nucleotides of the 3′-end trailer following the discriminator have the most pronounced effect on tRNase Z reaction, and the greatest anti-determinant effect was observed with the first two (–CC) (Mohan et al. 1999). Approximately 80 times more enzyme is required with mature tRNA (–CCA) (Fig. 7B) to achieve a processing time course comparable to that observed with natural sequence (Fig. 7A).
FIGURE 7.
Effect of the CCA anti-determinant on efficiency of tRNase Z reaction. T7 runoff templates were designed to produce a pre-tRNAHis substrate with the natural sequence A–AUG or with A–CCA of mature tRNA following the discriminator base (A–). Unlabeled pre-tRNAs were transcribed, gel-purified, treated with shrimp alkaline phosphatase, labeled with γ-32P-ATP using T4 polynucleotide kinase, and repurified (side designation S for substrate). These labeled pre-tRNAs were used in processing reactions sampled after 5, 10, and 15 min as indicated with vertical ticks below the product bands using enzyme concentrations of 0.0125 and 0.033 nM in the reactions with the –AUG pre-tRNA and with 0.0125, 0.033, 0.083, 0.25, 0.83, and 2.5 nM in reactions with –CCA mature tRNA as indicated below the gel. Comparable processing of –AUG substrate with 0.033 nM tRNase Z and of –CCA tRNA with 2.5 nM tRNase Z suggests that the CCA anti-determinant reduces reaction efficiency ∼80-fold.
The results of the relative efficiency experiments (Fig. 7) were confirmed and extended by performing kinetics with the pre-tRNA (3′-AUG) and mature tRNA (3′-CCA) substrates (Table 3). The kcat for 3′-AUG substrate is ∼4.5 times higher than that for tRNAHis with a 36-nt 3′-end trailer, and the kM is around two times higher for a combined slightly greater than twofold increase in kcat/kM (cf. Table 3, line 1, and Table 1, line 1). kcat for the –CCA substrate is reduced 80-fold relative to the –AUG substrate (the pertinent comparison) without an effect on kM. The CCA anti-determinant effect, an 80-fold reduction in kcat, thus arises from a component of fruit fly tRNase Z and does not require an ancillary factor.
TABLE 3.
tRNase Z processing kinetics with pre-tRNAHis-AUG and -CCA substrates
DISCUSSION
Five regions of pre-tRNA involved in recognition and catalysis by tRNase Z
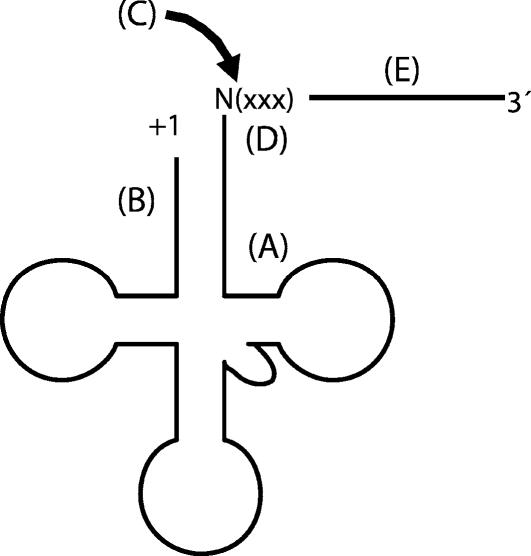
The pre-tRNA can be divided into five regions that contribute to tRNase Z binding and catalysis: the T arm, the acceptor stem, the scissile bond, the three nucleotides following the discriminator, and the rest of the 3′-end trailer (designated A–E, respectively, in Fig. 8). According to this model, the PxKxRN loop and Motif I, which are principally involved in CCA anti-determination and acceptor stem binding (through regions D and B in the substrate, respectively), are located close to each other and to the active site, and thus close to the scissile bond of the substrate (C). Structural and mutational data suggest that the T arm of the tRNA substrate (A) binds to the external binding domain of the enzyme while the 3′-end trailer (E) extends beyond the active site in an RNA exit channel. The significant contribution of Motif I to binding the acceptor stem, as well as the distribution of surface charge along the enzyme, suggest that much of the tRNA substrate specificity resides near the active site, with the external binding domain and exit channel providing additional stability.
FIGURE 8.
A pre-tRNA can be divided into five parts, each of them expected to contribute differently to tRNase Z recognition and catalysis. (A) The T arm; (B) acceptor stem; (C) tRNase Z processing site; (D) three nucleotides following the cleavage site (xxx = CCA of mature tRNA or “not-CCA” of a pre-tRNA); (E) additional sequence of the 3′-end trailer.
Substitutions in Motif II that reduce kcat by as much as 7500-fold have no effect on kM (Zareen et al. 2005); this region is thus concerned only with catalysis and the substrate recognition and binding components must reside elsewhere. The tRNase Z structure (de la Sierra-Gallay et al. 2005; Ishii et al. 2005; Kostelecky et al. 2006) and analysis of multiple sequence alignments and deletions (Schilling et al. 2005) suggest that an external binding domain remote from the catalytic core of tRNase Z is concerned with pre-tRNA recognition and binding. The minimal substrate for enzymes involved with pre-tRNA metabolism (RNase P, tRNase Z, CCA-adding enzyme) is a minihelix consisting of the coaxially stacked acceptor stem and T arm (McClain et al. 1987; Levinger et al. 1998; Shi et al. 1998). A specific interaction can therefore be proposed between the external binding domain of tRNase Z (Schilling et al. 2005) and the T arm of tRNA (A in Fig. 8), which are distal to the active site of the enzyme and the scissile bond of the substrate, respectively.
Significant increases in kM arise from substitutions in the PxKxRN loop (G438, C442, K446) (Fig. 5A) and Motif I region (D466 and especially R477) (Fig. 6A), which are too close to the active site to interact with the T arm of the substrate (Fig. 2). Effects of acceptor stem substitutions in the pre-tRNA substrate on tRNase Z reaction efficiency and kinetics have been previously reported (Marchfelder et al. 1996; Mohan and Levinger 2000; Levinger et al. 2004a; Yan et al. 2006), suggesting that a component of the enzyme could clamp the acceptor stem (B in Fig. 8).
Involvement of tRNase Z with the 3′-end trailer could have two components: a CCA anti-determinant domain, which distinguishes between mature tRNA with –CCA at the 3′ end and pre-tRNAs with 3′-end trailers possessing a sequence other than CCA following the discriminator (Fig. 8D), and a clamp for the 3′-end trailer (E in Fig. 8) which then productively binds proper substrates, stabilizing the scissile bond in the active site. A narrow exit channel was posited in interpretation of the B. subtilis model (de la Sierra-Gallay et al. 2005), suggesting a binding domain for the 3′-end trailer. Early support for such an exit channel was provided by analysis of pre-tRNAs with transcriptionally embedded CCA (Nashimoto 1997), in which increasing the length of the 3′-end trailer partially rescues mammalian tRNase Z catalysis.
Function of the PxKxRN loop is dominated by glycine at the amino boundary (G438) and the internal arginine (R448)
The great effect of the G438 substitution (a modest increase in kM and a much larger reduction in kcat combine to reduce kcat/kM 5000-fold) (Table 1; Fig. 5) suggests that the function of the entire loop can be ruined by replacement of this residue at the β1-loop boundary by allowing the secondary structure to extend beyond its natural end, drastically affecting shape and function of the PxKxRN loop. Replacement of P444 in mid-loop has comparatively little effect; P444 thus apparently does not contribute substantially to function of the PxKxRN loop, although it is conserved (Fig. 1A) and introduces a rigid bend (Fig. 2; de la Sierra-Gallay et al. 2005).
While it would be premature to offer a detailed model for regulation of tRNase Z activity, the conserved basic residues K446 and R448 may conduct surveillance on C74 and C75 as part of a CCA anti-determinant, interacting with region D in Figure 8. Analysis of processing kinetics using tRNA-C, -CC and -CCA showed that C75 (the second C) combined with C74 makes a greater contribution than C74 alone to the CCA anti-determinant (Mohan et al. 1999), just as substitution of R448 more strongly reduces tRNase Z reaction efficiency than substitution of K446.
The Motif I region consists of four structural elements
Motif I consists of a single aspartate that is important for tRNase Z catalysis (D25 in T. martitima, D37 in B. subtilis, and D466 in D. melanogaster tRNase Z) (Minagawa et al. 2004; de la Sierra-Gallay et al. 2005; Ishii et al. 2005), but the region subjected to detailed mutagenesis analysis was extended in both directions to include β3, the β3–α1 loop, α1, and part of the α1–β4 loop. Troughs are observed in the effects of substitutions on kcat and kcat/kM coinciding with β3 and α1 in the Motif I region except for Q474 (Fig. 6B,C), suggesting that Ala substitutions at the boundaries and in connecting loops usually affect tRNase Z function more than those within these structural elements.
Replacement of Motif I D466 with Ala reduces catalytic efficiency (kcat/kM) 3750-fold due to a 7.5-fold increase in kM and a 500-fold reduction in kcat relative to wild type. Substitutions in G468 and E469 close to the loop-α1 boundary also affect tRNase Z catalysis. Being strongly conserved (Fig. 1B), these residues might have both structural and electrostatic importance.
Y472 and Q474 correspond in position with two residues in α1 that were suggested to be involved in determination of the unusual cleavage site of T. maritima tRNase Z (Minagawa et al. 2004). Substitution of Y472 has practically no effect and Q474A has a moderate effect on both kM and kcat, which combine to make it one of the most important residues in this region. Neither substitution affects the tRNase Z cleavage site in fruit fly tRNAHis (data not shown).
R477 is located at the opposite end of the Motif I region from D466 (the loop on the carboxy side of α1) (Fig. 2C). Spikes in kM arise from the D466 and R477 substitutions (Fig. 5A), while substitution of R477 has an even greater effect on efficiency than D466. While D466 has previously documented effects on catalysis (the equivalent conserved residue in T. maritima tRNase ZS) (Minagawa et al. 2004), R477 must function differently, perhaps in binding the acceptor stem.
Effects of CCA on tRNase Z reaction
The CCA anti-determinant causes kcat to decrease 80-fold without affecting kM (Table 3). In earlier experiments with C–C–A added sequentially to the discriminator using unpurified fruit fly tRNase Z and tRNase Z purified from pig liver, decreases in kcat and increases in both kM and ki contributed to the CCA anti-determinant effect (Mohan et al. 1999). The discrepancy between present and earlier results could be explained if ancillary factors present in extracts but absent from expressed, affinity-purified tRNase Z can bind tRNA, affecting kM and ki for tRNase Z.
Since part of the CCA anti-determinant function may reside in the PxKxRN loop, its absence from T. maritima tRNase Z (Fig. 1A) could explain how this enzyme interacts atypically with CCA-containing pre-tRNAs after CCA (Minagawa et al. 2004; de la Sierra-Gallay et al. 2005). Interestingly, the first tRNase Z expressed and characterized (trz1 from A. thaliana, a tRNase ZS) (Schiffer et al. 2002) also lacks a PxKxRN loop, which could be why it lacks a tRNase Z anti-determinant (Schiffer et al. 2003). A. thaliana tRNase ZLs, on the other hand, have a PxKxRN loop (Fig. 1A). M. jannaschii tRNase Z has a PxKxRN loop so the lack of a CCA anti-determinant effect with this enzyme (Schiffer et al. 2003) must have a different explanation. tRNase Z processing experiments performed elsewhere typically use a 1000- to 100,000-fold higher enzyme concentration than that used here. The differential CCA effect (Fig. 7) can be reduced or even eliminated at a high enzyme concentration (data not shown), which could explain why no effect of –CCA was observed with M. jannaschii tRNase Z.
Characteristics of tRNase Z distinguish it from other members of the β-lactamase family that have been suggested to be endoribonucleases
Important endoribonucleases that for many years eluded identification may be cousins of tRNase Z in the β-lactamase family, including CPSF73 (implicated in pre-mRNA, including histone pre-mRNA, 3′ end processing) (Ryan et al. 2004; Dominski et al. 2005a) and the CPSF73-like integrator protein (suggested to be involved in snRNA 3′ end maturation) (Baillat et al. 2005; Dominski et al. 2005b). These proteins only function as site-specific endonucleases in concert with large macromolecular assemblies consisting of many additional protein subunits, while tRNase Z is apparently a soloist. Distinguishing features of tRNase Z include the external substrate recognition and binding domain (in the amino half of tRNase ZL), the PxKxRN loop, and Motifs I–V, which directly recognize the mutually complementary structural landmarks common to tRNAs (Fig. 8). Only weak homologies are observed between tRNase Z and its cousins outside of the His domain (Motif II); identification of the substrate recognition in the case of the other putative endoribonucleases thus requires more complex RNA–protein interactions.
Future prospects
Since the substitutions in the PxKxRN loop and Motif I region reveal residues that unexpectedly contribute to the catalytic efficiency of tRNase Z, it will be of interest to apply this scanning method to the homology blocks on the carboxy side of Motif II (e.g., Motifs III–V) and to the external recognition and binding domain in the amino half of tRNase ZL. Additionally, the ∼10-fold increases in kM arising from some of the single residue substitutions suggest direct contacts between enzyme and substrate that could be identified (as in Kühn-Hölsken et al. 2005). To finally understand the CCA anti-determinant it will be critical to solve the structures of tRNA–tRNase Z complexes.
MATERIALS AND METHODS
Construction, expression, and affinity purification of tRNase Z variants
Procedures for site-specific mutagenesis, baculovirus expression, and affinity purification of fruit fly tRNase Z and the methods for performing processing kinetics were recently described in detail in Zareen et al. (2005) and will therefore only be briefly outlined here. The fruit fly tRNase Z was designed to express from an internal methionine (M24, the first four encoded residues being MAAT) that is believed to be residue 1 of the nuclear form (Dubrovsky et al. 2004). Motif I mutations were constructed using the unique natural SacI site and the constructed XhoI site at the vector/insert boundary just beyond the stop codon, as previously described in Zareen et al. (2005). Because the SacI subcloning site (nt 1529) falls within the sequence that encodes the PxKxRN loop, these mutations were constructed using unique natural internal PflmI and Bpu10I sites (at nt 1146 and 1864, respectively), producing a 718-bp amplified DNA segment for subcloning. All variant DNA constructs were confirmed by sequencing (Herbert Irving Comprehensive Cancer Center, Columbia College of Physicians and Surgeons).
Templates for T7 RNA polymerase transcription
Runoff transcription with T7 RNA polymerase using plasmid templates has been described before; tRNAHis with a 36-nt 3′-end trailer and mature tRNAHis-CCA were designed to run off using DraI and NsiI RE ends, respectively, for use as tRNase Z substrates (Levinger et al. 1995; Mohan et al. 1999). The new template for tRNAHis-AUG (a positive control for tRNase Z reaction designed to closely match the length of mature tRNAHis) was prepared by introducing a SacI site at the appropriate position by amplification using a mismatched 3′-end primer and a universal 5′-end primer. tRNAHis with a long 3′-end trailer was either internally labeled during T7 transciption or unlabeled. Unlabeled tRNAHis-CCA and –AUG were transcribed and gel-purified, and a small portion of each transcript was 5′ end labeled by dephosphorylation using shrimp alkaline phosphatase followed by treatment with polynucleotide kinase and γ-32P-ATP and gel-purified again.
The 3′-AUG substrate appears to be 1 nt longer than the 3′-CCA substrate (Fig. 6A,B), probably due to runoff transcription of cloned templates using different RE ends (–AUG: SacI and –CCA: Nsi I, respectively). T7 RNA polymerase is prone to incorporate an unencoded nucleotide at the 3′ end (Milligan et al. 1987), perhaps depending on the choice of runoff RE and the encoded 3′-terminal sequence.
tRNase Z processing kinetics
Processing efficiencies were determined using labeled pre-tRNAHis with a 36-nt 3′-end trailer at a concentration of ∼10−10 M, more than 2 orders of magnitude lower than the lowest kM observed, so as to approximate zero-order kinetics, in which the % product/minute of reaction (V/[S]) evaluated early in a reaction time course is approximately proportional to V. For kinetic experiments, the concentration of labeled substrate was constant and total concentration of substrate was established by adding unlabeled pre-tRNAHis. While % product/minute (V/[S]) decreases with increasing [S], V (obtained by multiplying V/[S] by [S]) increases with [S]. The concentration of enzyme was adjusted as necessary based on results of efficiency experiments performed without added unlabeled substrate, and the substrate concentration range was adjusted so as to put the kM in the middle of the [S] range for each tRNase Z variant. Reliable, reproducible kinetic parameters were obtained with the lowest possible concentration of each variant enzyme. All variant enzymes were used for at least two kinetic experiments, and the reaction kinetics with the wild-type enzyme was performed alongside the variants every time a kinetic experiment was done. Comparisons to determine relative kcat/kM were made between variant and wild type performed on the same day.
Multiple sequence alignments
Multiple sequence alignments were prepared using ClustalW and displayed using GeneDoc.
Modeling the structure of the PxKxRN loop and Motif I in relation to Motif II
The tRNase Z structure model (de la Sierra-Gallay et al. 2005; PDB ID 1Y44) was visualized using New Cartoon in VMD with basic residues colored blue, acidic residues red, and neutral polar residues green.
ACKNOWLEDGMENTS
We thank Angelo Rossi for help with display of structure models and Zbigniew Dominski (University of North Carolina—Chapel Hill) for helpful discussions. This project was supported by grants from the NIH (S06GM08153) and the PSC-CUNY.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.4206.
REFERENCES
- Aebi M., Kirchner G., Chen J.Y., Vijayraghavan U., Jacobson A., Martin N.C., Abelson J. Isolation of a temperature-sensitive mutant with an altered tRNA nucleotidyltransferase and cloning of the gene encoding tRNA nucleotidyltransferase in the yeast Saccharomyces cerevisiae . J. Biol. Chem. 1990;265:16216–16220. [PubMed] [Google Scholar]
- Baillat D., Hakimi M.A., Naar A.M., Shilatifard A., Cooch N., Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123:265–276. doi: 10.1016/j.cell.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Castaño J.G., Tobian J.A., Zasloff M. Purification and characterization of an endonuclease from Xenopus laevis ovaries which accurately processes the 3′ terminus of human pre-tRNA-Met(i) (3′ pre-tRNase) J. Biol. Chem. 1985;260:9002–9008. [PubMed] [Google Scholar]
- Chen J.Y., Martin N.C. Biosynthesis of tRNA in yeast mitochondria. An endonuclease is responsible for the 3′-processing of tRNA precursors. J. Biol. Chem. 1988;263:13677–13682. [PubMed] [Google Scholar]
- de la Sierra-Gallay I.L., Pellegrini O., Condon C. Structural basis for substrate binding, cleavage and allostery in the tRNA maturase RNase Z. Nature. 2005;433:657–661. doi: 10.1038/nature03284. [DOI] [PubMed] [Google Scholar]
- Dominski Z., Yang X.C., Marzluff W.F. The polyadenylation factor CPSF-73 is involved in histone-pre-mRNA processing. Cell. 2005a;123:37–48. doi: 10.1016/j.cell.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Dominski Z., Yang X.C., Purdy M., Wagner E.J., Marzluff W.F. A CPSF-73 homologue is required for cell cycle progression but not cell growth and interacts with a protein having features of CPSF-100. Mol. Cell. Biol. 2005b;25:1489–1500. doi: 10.1128/MCB.25.4.1489-1500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky E.B., Dubrovskaya V.A., Levinger L., Schiffer S., Marchfelder A. Drosophila RNase Z processes mitochondrial and nuclear pre-tRNA 3′ ends in vivo . Nucleic Acids Res. 2004;32:255–262. doi: 10.1093/nar/gkh182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frendewey D., Dingermann T., Cooley L., Söll D. Processing of precursor tRNAs in Drosophila. Processing of the 3′ end involves an endonucleolytic cleavage and occurs after 5′ end maturation. J. Biol. Chem. 1985;260:449–454. [PubMed] [Google Scholar]
- Ishii R., Minagawa A., Takaku H., Takagi M., Nashimoto M., Yokoyama S. Crystal structure of the tRNA 3′ processing endoribonuclease tRNase Z from Thermotoga maritima . J. Biol. Chem. 2005;280:14138–14144. doi: 10.1074/jbc.M500355200. [DOI] [PubMed] [Google Scholar]
- Kostelecky B., Pohl E., Vogel A., Schilling O., Meyer-Klaucke W. The crystal structure of the zinc phosphodiesterase from Escherichia coli provides insight into function and cooperativity of tRNase Z family proteins. J. Bacteriol. 2006;188:1607–1614. doi: 10.1128/JB.188.4.1607-1614.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn-Hölsken E., Lenz C., Sander B., Luhrmann R., Urlaub H. Complete MALDI-ToF MS analysis of cross-linked peptide-RNA oligonucleotides derived from nonlabeled UV-irradiated ribonucleoprotein particles. RNA. 2005;11:1915–1930. doi: 10.1261/rna.2176605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinger L., Vasisht V., Greene V., Bourne R., Birk A., Kolla S. Sequence and structure requirements for Drosophila tRNA 5′ and 3′ end processing. J. Biol. Chem. 1995;270:18903–18909. doi: 10.1074/jbc.270.32.18903. [DOI] [PubMed] [Google Scholar]
- Levinger L., Bourne R., Kolla S., Cylin E., Russell K., Wang X., Mohan A. Matrices of paired substitutions show the effects of tRNA D/T loop sequence on Drosophila RNase P and 3′-tRNase processing. J. Biol. Chem. 1998;273:1015–1025. doi: 10.1074/jbc.273.2.1015. [DOI] [PubMed] [Google Scholar]
- Levinger L., Mörl M., Florentz C. A pathogenesis-associated mutation in human mitochondrial tRNALeu(UUR) leads to reduced 3′-end processing and CCA addition. J. Mol. Biol. 2004a;337:535–544. doi: 10.1016/j.jmb.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Levinger L., Mörl M., Florentz C. Mitochondrial tRNA 3′ end metabolism and human disease. Nucleic Acids Res. 2004b;32:5430–5441. doi: 10.1093/nar/gkh884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Deutscher M.P. Maturation pathways for E. coli tRNA precursors: A random multienzyme process in vivo. Cell. 1996;86:503–512. doi: 10.1016/s0092-8674(00)80123-3. [DOI] [PubMed] [Google Scholar]
- Li Z., Deutscher M.P. RNase E plays an essential role in the maturation of Escherichia coli tRNA precursors. RNA. 2002;8:97–109. doi: 10.1017/s1355838202014929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchfelder A., Schuster W., Brennicke A. In vitro processing of mitochondrial and plastid derived tRNA precursors in a plant mitochondrial extract. Nucleic Acids Res. 1990;18:1401–1406. doi: 10.1093/nar/18.6.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchfelder A., Brennicke A., Binder S. RNA editing is required for efficient excision of tRNAPhe from precursors in plant mitochondria. J. Biol. Chem. 1996;271:1898–1903. doi: 10.1074/jbc.271.4.1898. [DOI] [PubMed] [Google Scholar]
- McClain W.H., Guerrier-Takada C., Altman S. Model substrates for an RNA enzyme. Science. 1987;238:527–530. doi: 10.1126/science.2443980. [DOI] [PubMed] [Google Scholar]
- Milligan J.F., Groebe D.R., Witherell G.W., Uhlenbeck O.C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minagawa A., Takaku H., Takagi M., Nashimoto M. A novel endonucleolytic mechanism to generate the CCA 3′ termini of tRNA molecules in Thermotoga maritima . J. Biol. Chem. 2004;279:15688–15697. doi: 10.1074/jbc.M313951200. [DOI] [PubMed] [Google Scholar]
- Mohan A., Levinger L. The effects of matrices of paired substitutions in mid-acceptor stem on Drosophila tRNAHisstructure and end-processing. J. Mol. Biol. 2000;303:605–616. doi: 10.1006/jmbi.2000.4162. [DOI] [PubMed] [Google Scholar]
- Mohan A., Whyte S., Wang X., Nashimoto M., Levinger L. The 3′ end CCA of mature tRNA is an anti-determinant for eukaryotic 3′-tRNase. RNA. 1999;5:245–256. doi: 10.1017/s1355838299981256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mörl M., Marchfelder A. The final cut. The importance of tRNA 3′-processing. EMBO Rep. 2001;2:17–20. doi: 10.1093/embo-reports/kve006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashimoto M. Characterization of the spermidine-dependent, sequence-specific endoribonuclease that requires transfer RNA for its activity. Nucleic Acids Res. 1992;20:3737–3742. doi: 10.1093/nar/20.14.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashimoto M. Distribution of both length and 5′ terminal nucleotides of mammalian pre-tRNA 3′ trailers reflect properties of 3′ processing endoribonuclease. Nucleic Acids Res. 1997;25:1148–1154. doi: 10.1093/nar/25.6.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ow M.C., Kushner S.R. Initiation of tRNA maturation by RNase E is essential for cell viability in E. coli . Genes & Dev. 2002;16:1102–1115. doi: 10.1101/gad.983502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini O., Nezzar J., Marchfelder A., Putzer H., Condon C. Endonucleolytic processing of CCA-less tRNA precursors by RNase Z in Bacillus subtilis . EMBO J. 2003;22:4534–4543. doi: 10.1093/emboj/cdg435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K., Calvo O., Manley J.L. Evidence that polyadenylation factor CPSF-73 is the mRNA 3′ processing endonuclease. RNA. 2004;10:565–573. doi: 10.1261/rna.5214404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffer S., Rosch S., Marchfelder A. Assigning a function to a conserved group of proteins: The tRNA 3′-processing enzymes. EMBO J. 2002;21:2769–2777. doi: 10.1093/emboj/21.11.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffer S., Rosch S., Marchfelder A. Recombinant RNase Z does not recognize CCA as part of the tRNA and its cleavage efficiency is influenced by acceptor stem length. Biol. Chem. 2003;384:333–342. doi: 10.1515/BC.2003.039. [DOI] [PubMed] [Google Scholar]
- Schilling O., Späth B., Kostelecky B., Marchfelder A., Meyer-Klaucke W., Vogel A. Exosite modules guide substrate recognition in the ZiPD/ElaC protein family. J. Biol. Chem. 2005;280:17857–17862. doi: 10.1074/jbc.M500591200. [DOI] [PubMed] [Google Scholar]
- Shi P.Y., Weiner A.M., Maizels N. A top-half tDNA minihelix is a good substrate for the eubacterial CCA-adding enzyme. RNA. 1998;4:276–284. [PMC free article] [PubMed] [Google Scholar]
- Spath B., Kirchner S., Vogel A., Schubert S., Meinlschmidt P., Aymanns S., Nezzar J., Marchfelder A. Analysis of the functional modules of the tRNA 3′ endonuclease (tRNase Z) J. Biol. Chem. 2005;280:35440–35447. doi: 10.1074/jbc.M506418200. [DOI] [PubMed] [Google Scholar]
- Takaku H., Minagawa A., Takagi M., Nashimoto M. A candidate prostate cancer susceptibility gene encodes tRNA 3′ processing endoribonuclease. Nucleic Acids Res. 2003;31:2272–2278. doi: 10.1093/nar/gkg337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavtigian S.V., Simard J., Teng D.H.F., Abtin V., Baumgard M., Beck A., Camp N.J., Carillo A.R., Chen Y., Dayananth P., et al. A candidate prostate cancer susceptibility gene at chromosome 17p. Nat. Genet. 2001;27:172–180. doi: 10.1038/84808. [DOI] [PubMed] [Google Scholar]
- Vogel A., Schilling O., Spath B., Marchfelder A. The tRNase Z family of proteins: Physiological functions, substrate specificity and structural properties. Biol. Chem. 2005;386:1253–1264. doi: 10.1515/BC.2005.142. [DOI] [PubMed] [Google Scholar]
- Xiao S., Scott F., Fierke C.A., Engelke D.R. Eukaryotic ribonuclease P: A plurality of ribonucleoprotein enzymes. Annu. Rev. Biochem. 2002;71:165–189. doi: 10.1146/annurev.biochem.71.110601.135352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H., Zareen N., Levinger L. Naturally occurring mutations in human mitochondrial pre-tRNASer(UCN) can affect the tRNase Z cleavage site, processing kinetics and substrate secondary structure. J. Biol. Chem. 2006;281:3926–3935. doi: 10.1074/jbc.M509822200. [DOI] [PubMed] [Google Scholar]
- Zareen N., Yan H., Hopkinson A., Levinger L. Residues in the conserved His domain of fruit fly tRNase Z that function in catalysis are not involved in substrate recognition or binding. J. Mol. Biol. 2005;350:189–199. doi: 10.1016/j.jmb.2005.04.073. [DOI] [PubMed] [Google Scholar]
- Zhu L., Deutscher M.P. tRNA nucleotidyltransferase is not essential for Escherichia coli viability. EMBO J. 1987;6:2473–2477. doi: 10.1002/j.1460-2075.1987.tb02528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]