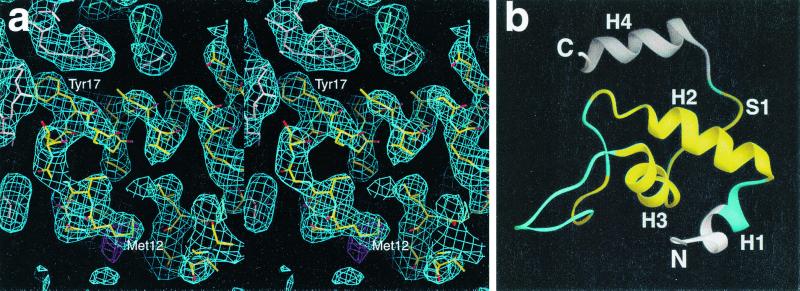
Figure 2.
Structure determination. (a) Stereo view of a portion of the 2|Fo| − |Fc| electron density map (3.2 Å, 1σ, shown in blue) calculated from the T. aquaticus core RNAP structure, showing a region corresponding to the ω subunit (including the N-terminal part of CR1 α-helix; at center, oriented horizontally) and nearby parts of β and β′. Atoms of ω are colored by atom type (C, yellow; O, red; N, blue; S, green). Atoms of β′ and β are colored pink and light blue, respectively. The SeMet difference Fourier peak (3σ) that corresponds to Met12 of ω is shown in magenta. Selected residues of ω are labeled. The figure was generated by using the program o (40). (b) Structure of the ω subunit in T. aquaticus RNAP core enzyme. A ribbon representation of T. aquaticus ω residues (residues 2–96) is shown. Residues of ω not included in the sequence alignment in Fig. 1 are illustrated in white; conserved regions CR1–CR3 are in yellow; nonconserved regions are in cyan. S1 is part of intersubunit β-sheet (two-strand antiparallel β-sheet with residues 1483–1487 of the C-terminal tail of β′).