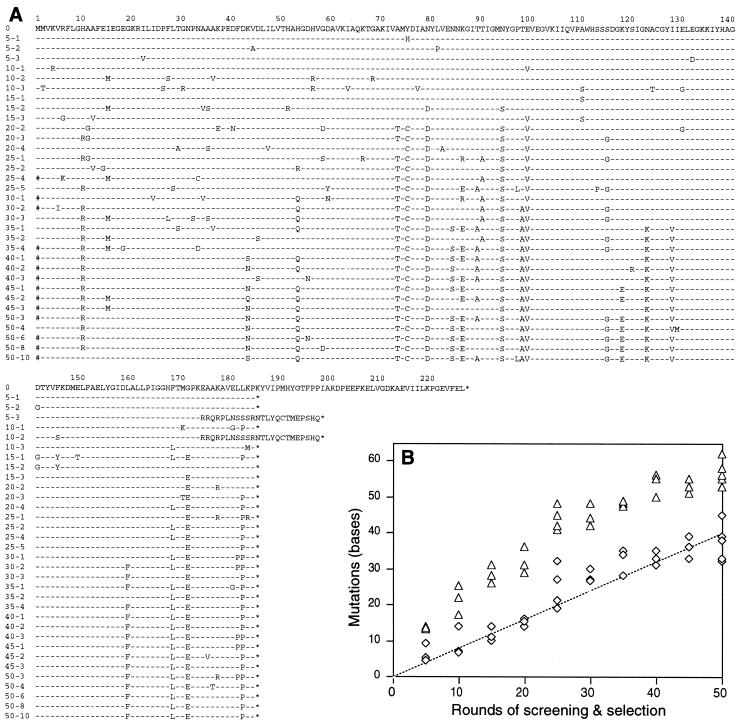
Figure 4.
Analysis of evolutionary intermediates. Entire evolved DNA fragments were sequenced for three clones at every five rounds. Four clones were sequenced for AR25 mutants. (A) Predicted amino acid substitutions in the coding region. Asterisks are stop codons and #, mutations in the original initiation methionine. (B) Each point represents the number of mutations found in a mutant. Mutations in the essential 3′ region (676–1,175 np) are triangles. The sum of silent mutations in the truncated coding region (121–675 np) and mutations in the 5′ region (1–120 np) is used as a molecular clock (diamonds). A line with a slope of 0.8 mutations/round is tentatively overlaid (see text).