Fig. 1.
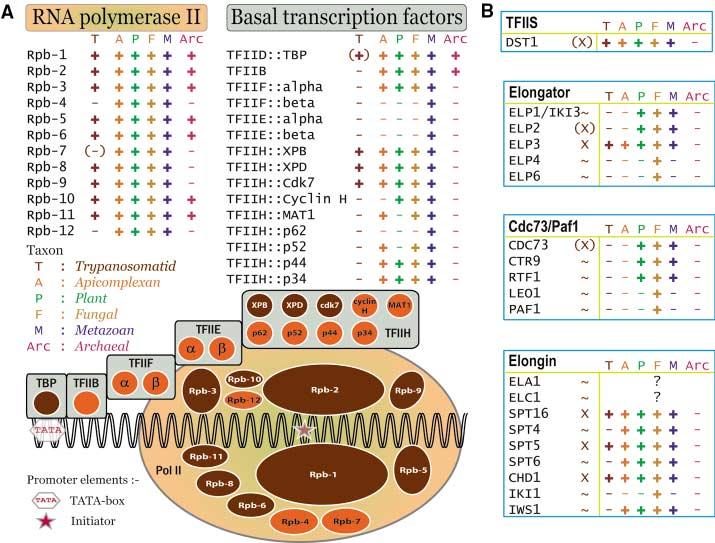
Trypanosomatid RNA polymerase subunits and transcription factors. (A) The TRIBE-MCL (table S2) protein families containing subunits of human RNA polymerase II (Rpb-1 to -12) and basal transcription factors (TBP and TFII-B, -E, -F, -H) are shown as ovals and circles respectively. Families containing Tritryp sequences are colored sepia and indicated by “+” or “(+)” (when the Tritryp gene is not a direct ortholog), whereas families lacking Tritryp sequences are indicated by “−” or “(−)” (when a Tritryp ortholog was detected only by BlastP analysis). Orthologs present in other taxa are indicated by “+”. The genomes queried for each taxon are detailed in (10). (B) Subunits of the yeast TFIIS, Elongator, Cdc73/Paf1, and Elongin complexes are displayed in blue boxes, with the first column of the row indicating whether Tritryp sequences with high “X”, weak “(X)”, or no “∼” similarity were detected. The remaining columns of the row are marked as in (A); “?” indicates that the S. cerevisiae sequence is not present in the TAP reference set.
