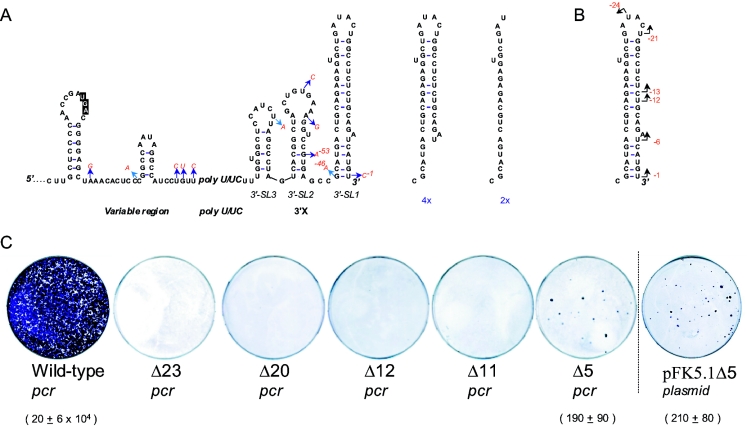
FIG. 1.
(A) Secondary structure of the 3′NTR and quasispecies therein derived from the serum of an HCV1b-infected patient. A total of 40 cDNA clones were sequenced: 29 cDNA clones corresponded to the WT sequence, and 11 individual cDNA clones contained single nucleotide changes indicated by arrows, followed by the nucleotide substitution (red). On the right side of the panel, four independent clones lacked five extreme 3′-end nucleotides, and two clones lacked 23 extreme 3′-end nucleotides. Note that the numbering starts at the last 3′ nucleotide, annotated with “−1,” and counts back. (B) The location of reported potential initiation sites is indicated by an arrow in SL1 (17, 28; the present study). (C) Effect of 3′-end deletions on cell colony formation using selectable replicons. WT is the pFK5.1 (Con1) construct amplified using T7 primer and reverse primer corresponding to the 3′ end (see the text for further details). Numbers below the plates refer to the CFU per microgram of in vitro-transcribed replicon RNA.