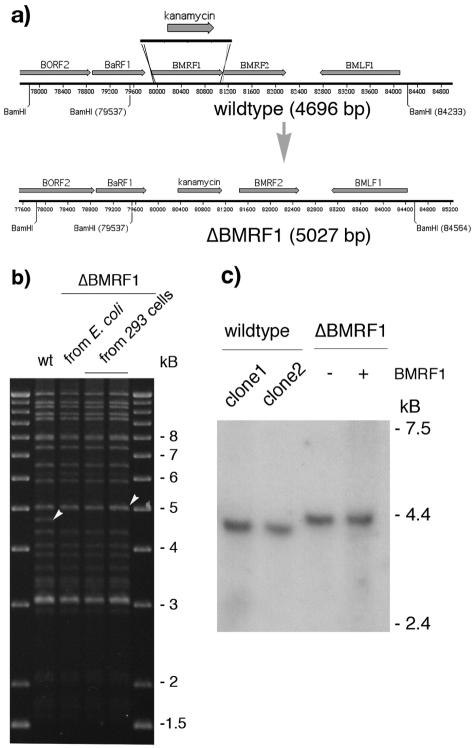
FIG. 1.
Schematic representation of the strategy used to construct a ΔBMRF1 EBV. (a) To disrupt the BMRF1 gene, the kanamycin resistance gene was amplified by PCR from pCP15 using primers that introduce 40-bp stretches of homology to sequences immediately upstream and downstream of the BMRF1 gene (EBV coordinates 79859 to 79899 and 81111 to 81150, respectively). Recombination of this linear DNA fragment with the wild-type EBV genome in E. coli leads to an exchange between the kanamycin cassette and the BMRF1 gene region, leading to an increase in size of the BamHI-M fragment from 4,696 bp to 5,027 bp. (b) Restriction analysis of the ΔBMRF1 mutant. Following recombination, the integrity of the recombinant ΔBMRF1 genome was analyzed at different steps of the mutant's construction. The mutant DNA was subjected to restriction analysis both before its transfer to the EBV-permissive 293 cell line and after its rescue from one of the obtained 293/ΔBMRF1 clones into E. coli cells. The restriction pattern obtained was compared to the one obtained after digestion of the wt EBV genome. Except for the wt 4,696-bp fragment that is exchanged for a 5,027-bp fragment in ΔBMRF1 (see white arrowheads), the overall structure of the mutant appears unchanged. This confirms the integrity and the mutant character of the 293/ΔBMRF1 cells. (c) Northern blot analysis of the transcription of the upstream BaRF1 gene of BMRF1. RNA from lytically induced cells was electrophoresed on a formaldehyde-1.2% agarose gel, blotted onto a Hybond N+ membrane, and hybridized with a radioactively labeled probe specific for the EBV BaRF1 gene. The intensities of the bands from two wild-type (B95.8) and noncomplemented or complemented ΔBMRF1 clones are very similar, proving that there is no effect of the BMRF1 deletion on the expression of the BaRF1 gene. The transcript in the ΔBMRF1 mutant is 330 bp longer since it spans the site of insertion of the kanamycin resistance gene.