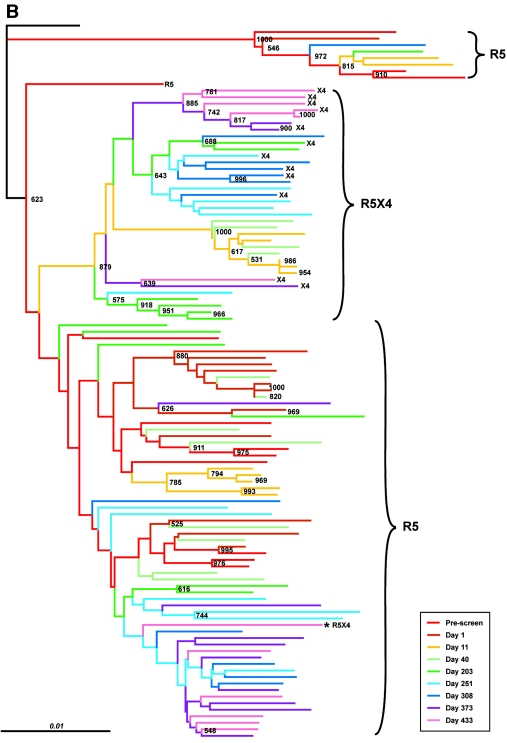
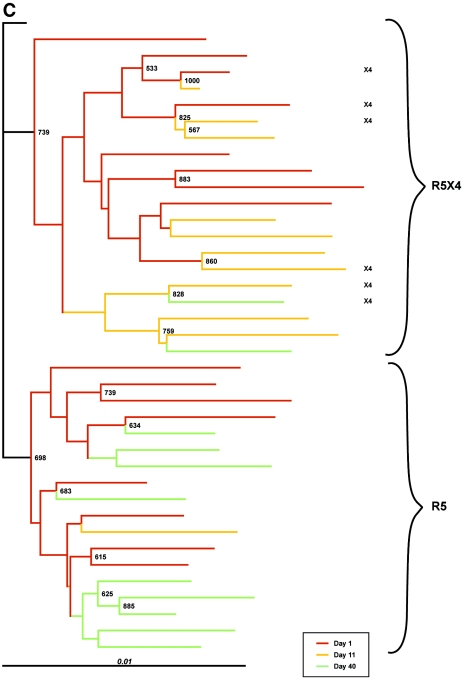
FIG.2.
CXCR4-using variants are genetically distinct from CCR5-tropic clones in each patient. Shown are the results of phylogenetic analysis of pre- and posttreatment gp160 sequences from patients A, B, and C (panels A, B, and C, respectively). Neighbor-joining trees were generated using the Clustal W program from an alignment edited to remove V4 and V5. The “disregard gapped” columns and “correct for multiple hits” options were selected, and a consensus sequence was generated from the earliest set of clones and used as an outgroup to root the tree. The numbers at the nodes are values determined from 1,000 bootstrap trials Sequence diversity is represented by horizontal distance. The phenotypic tropism assignment of the clones is indicated on each panel.