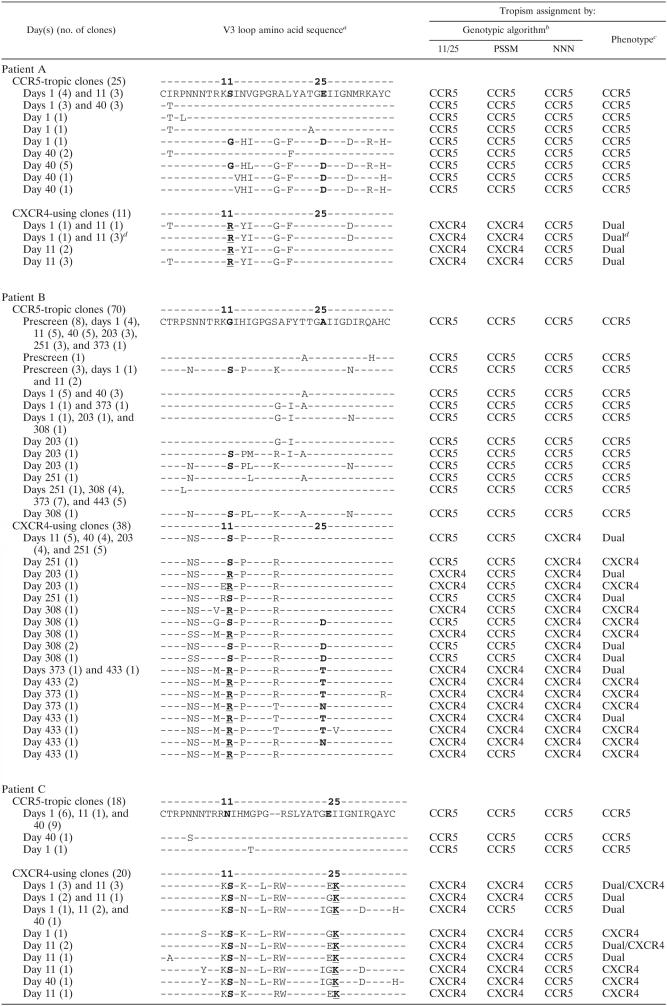
TABLE 3.
V3 loop sequences of functional Env clones
Amino acid positions 11 and 25 are in boldface, and amino acids at these positions associated with CXCR4 tropism are underlined. Amino acid changes relative to the earliest clone are shown. Amino acids associated with CXCR4 use are in boldface.
Determined using the 11/25 algorithm (8, 14, 20), the PSSM of Jensen et al. (21), or the presence/absence of a glycosylation site (NNN) (34). Boldface entries indicate instances where the genotypic call did not correlate with the assigned coreceptor phenotype.
Determined using the PhenoSense HIV Entry assay.
Virus derived from patient A, day 11, clone 10 clustered with dualtropic clones in the genotypic analysis but in the PhenoSense assay gave very low RLU readings only on CCR5-expressing cells.