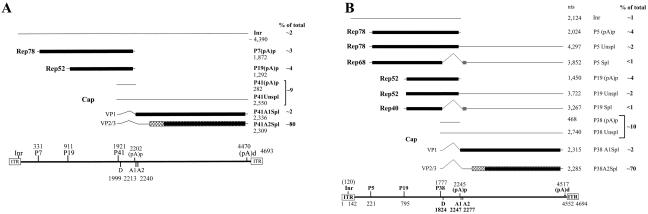
FIG. 4.
Transcription maps of B-AAV and A-AAV. (A) Transcription map of B-AAV. The genome of B-AAV is diagrammed, with transcription landmarks shown to scale, including the ITR, P7 promoter, P19 promoter, P41 promoter, splice donor (D), and acceptor (A1 and A2) sites, the internal polyadenylation site [(pA)p], and the distal polyadenylation site [(pA)d]. All of the RNA species detected in our assays are diagrammed with an indication of their identities and respective sizes. The only Rep-encoding transcripts generated by B-AAV were the Rep 78 and Rep 68 mRNAs, which were polyadenylated at the (pA)p site. RNA initiation within the ITR was from multiple start sites (data not shown). The approximate relative accumulated levels of each transcript type in total RNA is given as a percentage. (B) Transcription map of A-AAV. The genome of A-AAV is shown with transcription landmarks shown to scale, including the ITR, P5 promoter, P19 promoter, P38 promoter, splice donor (D) and acceptor (A1 and A2) sites, the internal polyadenylation site [(pA)p], and the distal polyadenylation site [(pA)d]. All of the RNA species are shown with their respective sizes. In contrast to B-AAV and AAV5, a significant percentage of P5- and P19-generated mRNAs read through the internal (pA)p site and are found either as unspliced (unspl) or spliced (spl) species. The approximate relative accumulated levels of each transcript type in total RNA as determined by RPAs is given as a percentage.