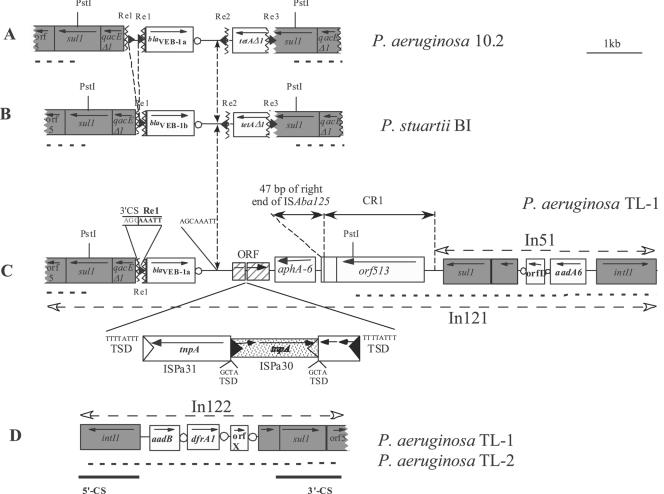
FIG. 1.
Schematic representations of the genetic environment of the blaVEB-1-like gene (A) in P. aeruginosa 10.2 (2) from India, (B) in P. stuartii BI (3) from Algeria, (C) in P. aeruginosa TL-1 from Bangladesh, and (D) of In122 found in P. aeruginosa TL-1 and TL-2. The Re are represented as black triangles, and the flanking sequence identity breakpoints in the blaVEB-1-like gene environment are designated by vertical broken lines. The coding regions are shown as boxes, with an arrow indicating the orientation of transcription and white circles indicating 59-be's. Dark-gray boxes correspond to genes or ORFs present on class 1 integron conserved sequences. Restriction sites that were used for cloning are indicated. Dashed lines in bold indicate regions that were identified using PCR (the sequences of primers are available upon request). Horizontal dashed-lined arrows indicate the homology breakpoints in In121, compared to the structures found in P. aeruginosa 10.2 and P. stuartii BI. Filled and empty triangles represent inverted repeats of insertion sequences. Target site duplications are also indicated.