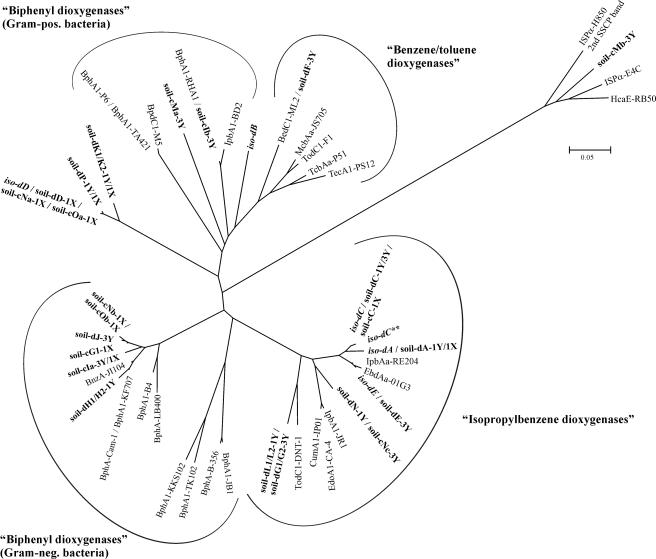
FIG. 3.
Phylogenetic analysis of deduced peptide sequences from ISPα gene segments recovered from SSCP profiles (comprising 158 to 161 amino acids, corresponding to positions 224 to 384 of BphA1 from B. xenovorans LB400). Characterized ISPα peptides of the toluene/biphenyl oxygenase subfamily (IpbAa, Pseudomonas putida RE204 [AF006691]; EbdAa, Pseudomonas putida 01G3 [AJ293587]; IpbA1, Pseudomonas sp. strain JR1 [U53507]; CumA1, Pseudomonas fluorescens IP01 [D37828]; EdoA1, Pseudomonas fluorescens CA-4 [AF049851]; BphA1, Burkholderia sp. strain JB1 [AJ010057]; BphA1, Comamonas testosteroni B-356 [U47637]; BphA1, Comamonas testosteroni TK102 [AB086835]; BphA1, Pseudomonas sp. strain KKS102 [D17319]; BphA1, Burkholderia xenovorans LB400 [M86348]; BphA1, Pseudomonas sp. strain B4 [U95054]; BphA1, Pseudomonas pseudoalcaligenes KF707 [M83673]; BphA, Pseudomonas sp. strain Cam-1 [AY027651]; BnzA, Pseudomonas aeruginosa JI104 [E04215]; BphA1, Rhodococcus erythropolis TA421 [D88020]; BphA1, Rhodococcus globerulus P6 [X80041]; BpdC1, Rhodococcus sp. strain M5 [U27591]; BphA1, Rhodococcus sp. strain RHA1 [D32142]; IpbA1, Rhodococcus erythropolis BD2 [U24277]; BedC1, Pseudomonas putida ML2 [AF148496]; McbAa, Ralstonia sp. strain JS705 [AJ006307]; TodC1, Pseudomonas putida F1 [J04996]; TcbAa, Pseudomonas sp. strain P51 [U15298]; TecA1, Ralstonia sp. strain PS12 [U78099]; ISPα, Variovorax paradoxus E4C [AF209469]; HcaE, Bordetella bronchiseptica RB50 [NC_002927]; ISPα, Cupriavidus necator H850 [2nd SSCP band]) and ISPα peptides from bacterial isolates (bold italics) and soil samples (bold) are shown. The ISPα peptides obtained from soil (soil-) were designated according to the procedure of sequence recovery, i.e., direct sequencing of reamplified SSCP products (d) or sequencing of cloned PCR-amplified products (c), followed by band position (A through P) and sample origin (1X, 1Y, or 3Y). In cases where more than one consensus sequence was retrieved by cloning, the different sequence types are indicated by lowercase letters (a, b, or c) after the band position designation. The ISPα peptide sequences from isolates (iso-) were deduced from directly sequenced reamplification products. The tree was constructed by using the neighbor-joining method in MEGA with the p-distance model and pairwise deletion of gaps/missing data. The scale bar indicates the percent divergence. The names of ISPα peptide clusters were assigned according to the native substrates used for isolation of their hosts and characterization of their activity. pos., positive; neg., negative.