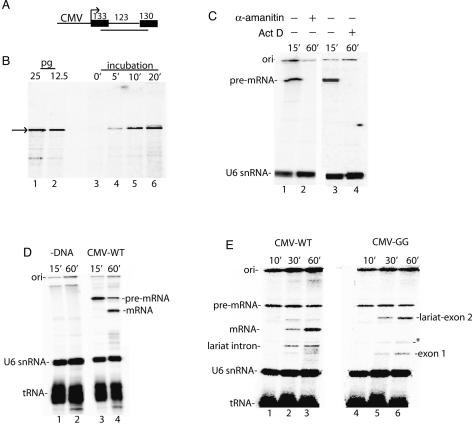
Figure 1.
RNAP II transcription and pre-mRNA splicing in vitro. (A) Structure of the DoA1 DNA template used to optimize transcription and splicing. The sizes of the exons and intron are indicated. The line below the DoA1 DNA template indicates the SP6 RNA probe, which protects 234 nt of T7-AdML and CMV-DoA1 (54 nt of exon 1, the entire intron, and 57 nt of exon 2). (B, lanes 1,2) RNase protection of known amounts of AdML T7 transcript. (Lanes 3–6) RNase protection of RNAP II transcript, which was generated by incubating the DoA1 DNA template in HeLa nuclear extract for the indicated times. The arrow indicates the protected transcript. (C, lanes 1,3) 32P-UTP and the CMV DNA template were incubated under transcription/splicing conditions for 15 min. α-Amanitin (lane 2) or Actinomycin D (lane 4) was added at the beginning of the transcription/splicing reaction and incubation was continued for 60 min. (D) 32P-UTP and the DoA1 DNA template were incubated under transcription/splicing conditions for 15 min (lane 3) or 60 min (lane 4). The DNA was omitted in the reactions shown in lanes 1 and 2. (E) Same as D, except that the CMV GG and wild-type (WT) DNA templates were compared. Asterisk indicates a nonspecific band. The endogenous U6 snRNA and tRNA are indicated. RNA was fractionated on an 8% denaturing polyacrylamide gel and detected by PhosphorImager.