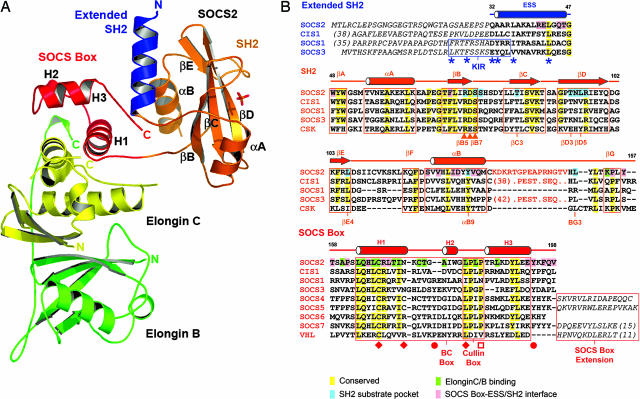
Fig. 1.
Overall structure of the SOCS2–elongin BC ternary complex and structure-based sequence alignment. (A) The different domains and complex components are differentiated by colors showing the SOCS2 SH2 domain in orange, the ESS in blue, the SOCS box in red, and two elongins in yellow and green. Secondary structure elements discussed in the Results are labeled. (B) Conserved and domain interface residues are highlighted by different colors in the sequence alignment. An unstructured 15-residue insertion in the SOCS2 BG loop is represented by orange text and is the site of predicted PEST sequences in CIS1 and SOCS3 (21). Additional N- and C-terminal extensions are defined by italic text. The positions of mutations that disrupt the SOCS1–JAK2 (22), SOCS2–GHR (11), VHL–elongin C (18), and SOCS1–Cul-5 (16) interactions are indicated by asterisks, triangles, diamonds, and open squares, respectively. Filled circles mark SOCS3 phosphorylation sites, which disrupt the SOCS3–elongin C interaction (20). The SOCS2 SH2 structure is most similar to the structures of C-terminal Src kinase (PDB ID code 1K9A) and Csk-homologous kinase (PDB ID code 1JWO).