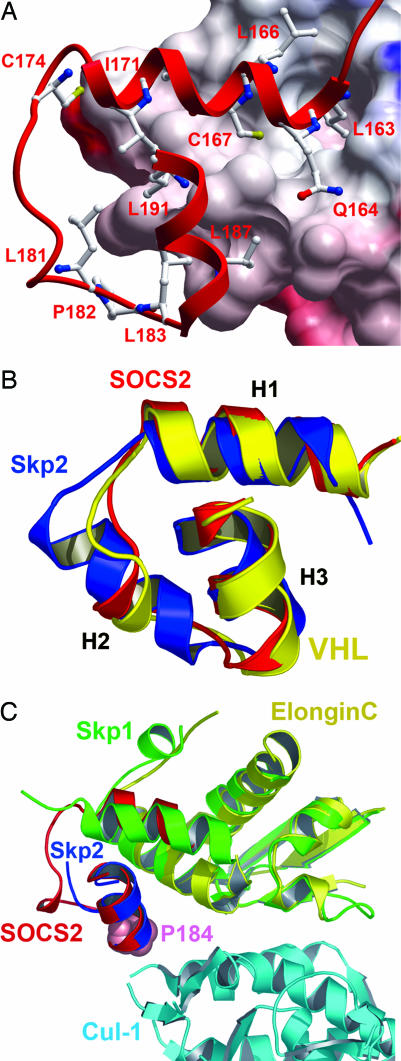
Fig. 2.
Comparative structural interactions and conservation of the SOCS box, BC box, and F box. (A) SOCS2 binds elongin C (electrostatic surface shown) in a hydrophobic interface of ≈2,200 Å2 (for clarity, elongin B is not shown). Deep pockets accommodate residues from SOCS2 H1 (L163 and C167). (B) The three core helices of the SOCS box (red) show a remarkable structural conservation with the VHL BC box (yellow) and Skp2 F box (blue), forming a common structural motif to link E3 substrate recognition domains with the ubiquitin ligase complex. (C) The crystal structure of the Skp2-Skp1–Cul-1–Rbx1 ternary complex provides a template for the study of SOCS box–elongin C binding to Cul-5 (38). Because of different packing arrangements in their respective complexes, Skp1 shows structural and functional similarity to elongin C and SOCS2 H1, whereas H1 from Skp2 provides equivalence to SOCS2 H3 [consistent with previous Skp2-Skp1 comparison with VHL (39)]. SOCS2 P184 (shown in space-fill) occurs at the likely Cul-5 interface and terminates the “LPXP” cullin box. Mutation of this position in SOCS1 determines Cul-5 interaction (16).