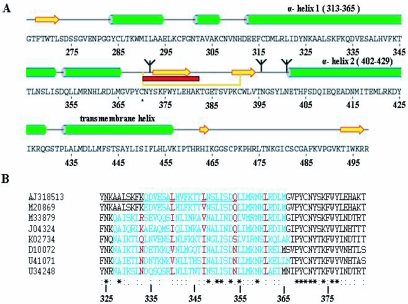
FIG. 1.
Secondary structure prediction of the GP-2 subunit and identification of a trimeric heptad repeat pattern. (A) Secondary structure prediction for the LCMV-WE GP-2 sequence (AJ318513) used in this study (34). Green cylinders represent predicted α-helices, and yellow arrows represent β-sheets. Three prominent α-helices were identified: α-helix 1 ranged from aa 313 to aa 365, and α-helix 2 ranged from aa 402 to aa 429, located just prior to the transmembrane helix (10). A predicted disulfide-bonded loop is indicated by a yellow line connecting cysteines 370 and 391. N-linked glycosylation sites are indicated by trees. The major antigenic site on GP-2 (aa 371 to aa 382), which is recognized by the antibody WE 83.4, is highlighted in red (54). (B) Results of the LearnCoil-VMF analysis for aligned sequences of LCMV-WE (AJ318513), LCMV-Armstrong (M20869), Mopeia (M33879), Lassa (J04324), Pichinde (K02734), Junín (D10072), Sabia (U41071), and Oliveros (U34248) viruses (48). Sequences containing a potential trimeric heptad repeat pattern are highlighted in blue. Amino acids in the α-position within the heptad repeat pattern are colored in red. The underlined stretch within the LCMV-WE sequence (AJ318513) matches the sequence of peptide 1, which was tested for its oligomerization potential. The degree of conservation among the aligned sequences is indicated at the bottom: an asterisk indicates complete conservation, and two dots and one dot indicate more-conservative mutations with respect to charge and size.