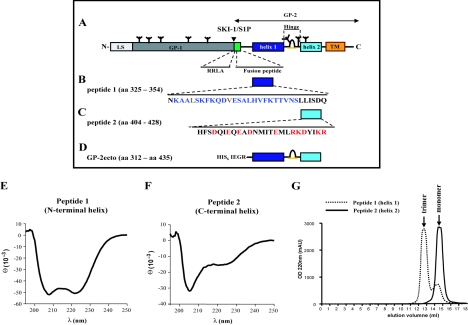
FIG. 2.
An α-helical peptide corresponding to the heptad repeat pattern containing helix 1 forms stable oligomers at pH 7.35, whereas an α-helical peptide corresponding to helix 2 has no potential to oligomerize. (A) Wild-type LCMV-WE GP sequence. At the N terminus, the leader sequence (LS) is indicated, followed by the receptor binding subunit GP-1 and the transmembrane subunit GP-2. N-linked carbohydrates are indicated by trees. The subtilase SKI-1/S1P cleavage site and the corresponding consensus sequence (RRLA) are marked by an arrowhead. Structural features within the GP-2 subunit are highlighted: the predicted N-terminal hydrophobic fusion peptide is colored in green (29, 30), the identified heptad repeat pattern containing α-helix 1 in dark blue, and α-helix 2 in light blue. Helices 1 and 2 are connected by a hypothetical disulfide-bonded loop (27). At the very C terminus, the transmembrane helix (TM) and the cytoplasmic tail are shown. (B and C) Full-length sequences of peptides used in this study. Amino acids in the peptide 1 sequence, which have been identified by the LearnCoil-VMF program, are colored in blue. Charged amino acids in the peptide 2 sequence are colored in red, thus emphasizing the strong amphipathic character of this helical region. (D) N-terminally tagged GP-2 ectodomain construct (GP-2ecto) expressed in E. coli. The construct contained a C316S point mutation to avoid artificial interchain disulfide bridges. (E and F) Circular dichroism spectra for peptide 1 and peptide 2, respectively. (G) Superdex 75 gel filtration chromatograms for peptide 1 (3.1 kDa) and peptide 2 (3.3 kDa) at pH 7.35. Peptide 1 shows a main peak at an elution volume of 13 ml and a minor shoulder peak at 15 ml, whereas peptide 2 shows a single peak at 15 ml. The main peak fractions of each peptide were concentrated and analyzed by dynamic light scattering. OD 220nm, optical density at 220 nm; mAU, milli-adsorption units.