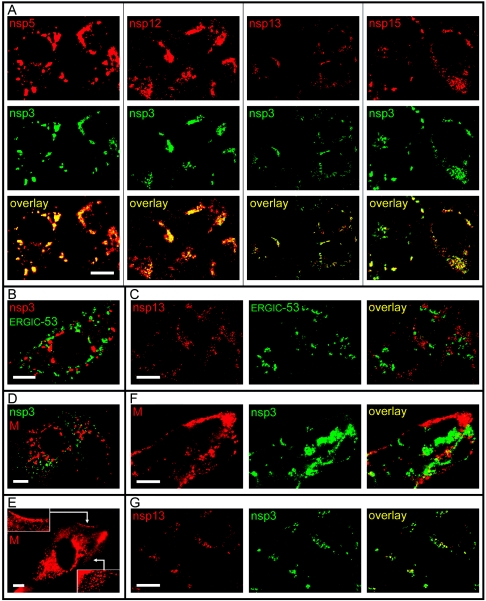
FIG. 3.
Confocal IF microscopy analysis of the intracellular distribution of various SARS-CoV replicase subunits in infected Vero E6 cells. (A) Double-labeling experiments (9 h p.i.) using an AF488-coupled IgG fraction purified from an anti-nsp3 serum and antisera recognizing nsp5, nsp12, nsp13, and nsp15. Extensive colocalization of these five nonstructural proteins was observed throughout infection. (B) Double-labeling experiment (9 h p.i.) for SARS-CoV nsp3 and the ERGIC-53 cellular marker protein. (C) Double-labeling experiment (18 h p.i.) for the SARS-CoV nsp13 helicase and the ERGIC-53 cellular marker protein, illustrating the complete separation of the nsp13 and the ERGIC at late time points in infection. (D) Double-labeling experiment (6 h p.i.) for SARS-CoV nsp3 and the viral M protein, which localizes to the Golgi complex at this time point. (E) Labeling for the SARS-CoV M protein at 9 h p.i., showing the spread of the protein throughout the cytoplasm, presumably due to the traffic of progeny virions towards the plasma membrane. Insets illustrate the strong labeling of the region just beneath the plasma membrane. (F) Double-labeling experiment (18 h p.i.) for SARS-CoV nsp3 and M protein, confirming the almost-complete separation of the two proteins also at late time points in infection. (G) Double-labeling experiment (18 h p.i.) using an AF488-coupled IgG fraction purified from an anti-nsp3 serum and an antiserum recognizing nsp13, illustrating the colocalization of the two proteins also at late stages of infection. In general, late in infection, the nsp13 signal was found to decline more rapidly than that of nsp3, suggesting differences in turnover of these two proteins. Bar, 10 μm.