Figure 1.
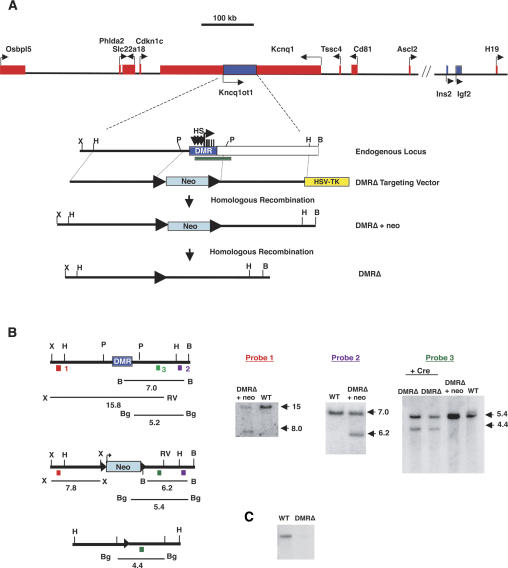
Deletion of the KvDMR. (A) The top line represents a physical map of the imprinted domain in mouse distal chromosome 7, with maternally expressed genes represented by red boxes and paternally expressed genes shown as blue boxes. Depicted below is an enlargement of the KvDMR (blue box), illustrating the three DNase I hypersensitivity sites (HS; vertical arrows), the Kcnq1ot1 transcription start site (horizontal arrow), and the five repeats (vertical bars). The location of the KvDMR deletion described by Fitzpatrick et al. (2002) is depicted by the green rectangle. The targeting vector is shown below, in which the neomycin resistance gene (neo) flanked by loxP sites (closed triangles) replaces 3.6 kb of the DMR. The herpes simplex virus thymidine kinase gene (HSV-TK) was included to provide negative selection. The structures of the targeted loci, before and after the deletion of the neo gene, are indicated. (B) Southern blot analysis of DNAs prepared from wild-type (WT), heterozygous DMRΔ +neo, and heterozygous DMRΔ (−neo) mice after digestion with restriction enzymes and hybridization with the probes 1–3 as indicated in the diagrams. The diagrams of the endogenous and targeted loci depict the positions and sizes of the fragments detected by the probes. (C) RNA protection analysis of RNAs isolated from wild type and mice inheriting DMRΔ paternally using probe 3. (P) PstI; (X) XhoI; (H) HindIII; (B) BamHI; (RV) EcoRV; (Bg) BglI.