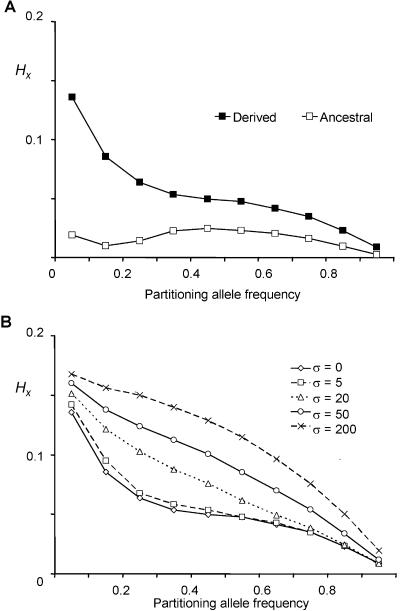
Figure 2.
Comparison of mean regional HX values for ancestral and derived alleles for simulated data sets, with the central target allele set to attain a range of frequencies. A, Ancestral and derived alleles modeled as neutral sites. B, Derived alleles under positive selection with different population-scaled selection coefficients (σ=2Ns, where 2N is the population size and s is the selection coefficient). Each simulation was run 1,000 times for a given allele frequency. SEMs were always <0.00045—well within the symbols shown. As expected, HX values decay with increasing allele frequency. Haplotype homozygosity was preserved on haplotypes marked by derived alleles and around alleles that have undergone rapid positive selection.