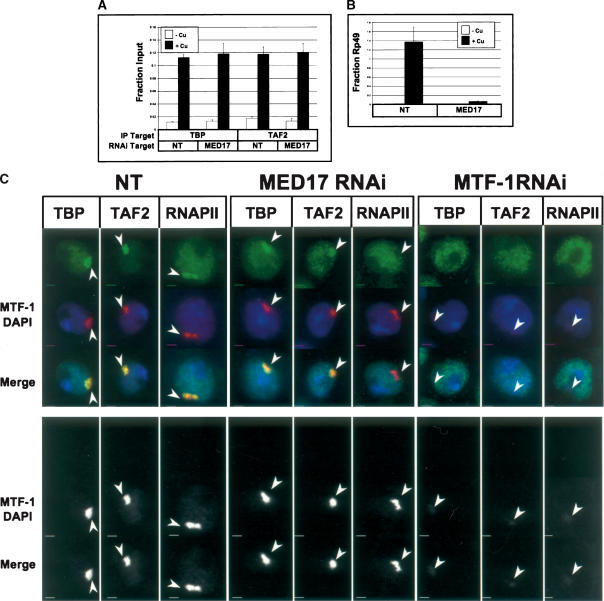
Figure 6.
Independent recruitment of TFIID to MtnA transgene by MTF-1. (A) ChIP analysis of TFIID recruitment in the absence of MED. S2 cells were treated with dsRNA as indicated at the left and subsequently immunoprecipitated with the antibody indicated below the graph. White bars represent results from untreated S2 cells, and black bars represent results from S2 cells treated with copper for 6 h at 25°C. (B) Quantitative PCR analysis of the MtnA mRNA, as in Figure 1B. (C) S2 cells harboring the MtnA transgene treated with dsRNAs against MED17 and MTF-1 were induced with CuSO4 for 2 h and were examined for TFIID and Pol II recruitment using anti-TBP, anti-TAF2, and anti-RPB2. Arrows indicate the location of the transgene marked by MTF-1. The bottom set of black and white panels shows the MTF-1 channel only to illustrate the residual amount of MTF-1 signal after RNAi treatment used to estimate the location of the transgene. Scale bar, 2 μm.