FIGURE 2.
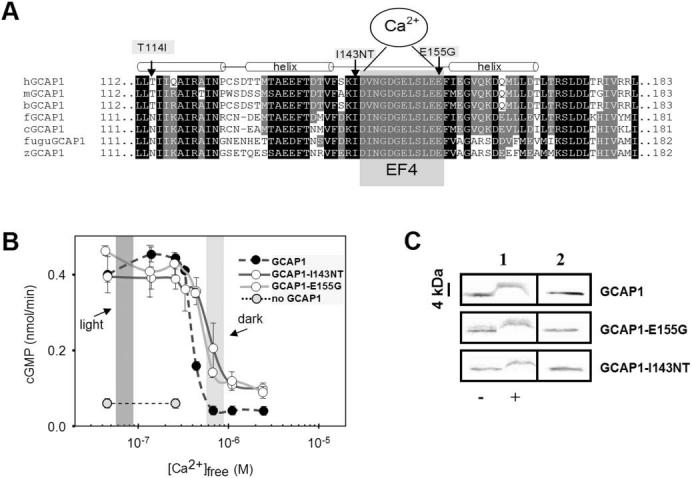
Amino acid sequence of vertebrate GCAP1s around EF4 and biochemical analysis of mutant GCAPs. (A) Sequence of vertebrate GCAP1s around the Ca2+-binding loop EF4. EF4 is highlighted by gray shading, surrounded by two α-helical structures (approximately three helical turns). The mutations changing the hydrophobic residue Ile143 and the acidic residue E155 are identified by arrows. A third mutation found in an independent family (Fig. 1B) is situated in the C-terminal helix of EF3. It is unclear whether this mutation causes retinal disease. (B) Stimulation of GC activity in ROS membranes by normal and mutant GCAPs as a function of Ca2+. The dark-shaded area indicates low [Ca2+]free expected in the light-adapted photoreceptors; the light-shaded area reflects high [Ca2+]free expected in the dark-adapted photoreceptors. Assays were carried out with addition of 3 μM GCAPs and were repeated at least three times. Error bars: SD for GC activity stimulated by GCAPs. (C) Stained SDS gel of GCAP1, GCAP1-I143NT, and GCAP1-E155G in the presence or absence of Ca2+ (column 1);-, absence of EGTA, +, presence of EGTA. Column 2 is a control immunoblot for GCAPs in water. GCAPs display a shift in mobility in the absence of bound Ca2+. The antibody used is UW14.21
