Abstract
The mitochondrial inner membrane contains two separate translocons: one required for the translocation of matrix-targeted proteins (the Tim23p-Tim17p complex) and one for the insertion of polytopic proteins into the mitochondrial inner membrane (the Tim54p-Tim22p complex). To identify new members of the Tim54p-Tim22p complex, we screened for high-copy suppressors of the temperature-sensitive tim54-1 mutant. We identified a new gene, TIM18, that encodes an integral protein of the inner membrane. The following genetic and biochemical observations suggest that the Tim18 protein is part of the Tim54p-Tim22p complex in the inner membrane: multiple copies of TIM18 suppress the tim54-1 growth defect; the tim18::HIS3 disruption is synthetically lethal with tim54-1; Tim54p and Tim22p can be coimmune precipitated with the Tim18 protein; and Tim18p, along with Tim54p and Tim22p, is detected in an ∼300-kDa complex after blue native electrophoresis. We propose that Tim18p is a new component of the Tim54p-Tim22p machinery that facilitates insertion of polytopic proteins into the mitochondrial inner membrane.
INTRODUCTION
Mitochondrial function requires the import of hundreds of proteins synthesized in the cytosol (for review, see Ryan and Jensen, 1995; Pfanner, 1998; Ryan et al., 1998; Rassow et al., 1999). In the yeast Saccharomyces cerevisiae, mitochondrial protein import uses multiple subunit translocases found in the mitochondrial outer membrane (called the TOM complex) and in the inner membrane (called the TIM complexes). Most proteins destined for mitochondria carry amino-terminal targeting signals called presequences. Presequences vary in length and primary amino acid sequence, yet they share a common motif of a positively charged amphipathic helix (Allison and Schatz, 1986; Roise et al., 1986, 1988; Roise, 1992). Once in the matrix, the presequence is removed by a two-subunit processing protease called MPP (McAda and Douglas, 1982; Yaffe et al., 1985; Jensen and Yaffe, 1988; Pollock et al., 1988; Witte et al., 1988; Yang et al., 1988). Some proteins destined for the mitochondrial inner membrane (IM) carry a cleavable presequence followed by one or more hydrophobic membrane-spanning segments (Stuart and Neupert, 1996). The transmembrane segments are proposed to either function as stop-transfer sequences in the IM (Miller and Cumsky, 1991, 1993) or facilitate the insertion of the polypeptide into the IM after its complete import into the matrix (Mahlke et al., 1990; Herrmann et al., 1997).
Mitochondrial precursor proteins bind to one of several receptors on the mitochondrial surface (Hase et al., 1983; Hines et al., 1990; Steger et al., 1990; Moczko et al., 1993; Ramage et al., 1993; Gratzer et al., 1995; Hönlinger et al., 1995; Lithgow et al., 1995; Moczko et al., 1997). Precursors are then translocated across the outer membrane (OM) via the TOM complex, which includes Tom40p, Tom22p, Tom7p, Tom6p, and Tom5p (Alconada et al., 1995; Honlinger et al., 1996; Dietmeier et al., 1997; Hill et al., 1998). The mitochondrial IM appears to have at least two separate import machines. One multiple subunit complex, the Tim23p-Tim17p translocon, is required to translocate proteins across the IM into the matrix (Emtage and Jensen, 1993; Maarse et al., 1994; Ryan et al., 1994; Lohret et al., 1997). Tim44p and mt-Hsp70 provide the pulling force for this import (Pfanner et al., 1994; Rassow et al., 1994; Stuart et al., 1994; Berthold et al., 1995; Glick, 1995). Precursors that carry positively charged presequences are targeted to Tim23p-Tim17p after their translocation across the OM (Moro et al., 1999). The Tim23p-Tim17p complex also plays a role in the insertion of some proteins into the IM.
A second translocase in the IM is the Tim54p-Tim22p complex (Sirrenberg et al., 1996; Kerscher et al., 1997). Tim54p and Tim22p are required for the insertion of many polytopic proteins into the IM. Substrates for the Tim54p-Tim22p complex do not carry cleavable, amino-terminal presequences. These include members of the mitochondrial carrier family, such as the phosphate carrier (PiC) and the ADP/ATP carrier, as well as the membrane components of the IM translocases, Tim23p, Tim22p, and Tim17p (Sirrenberg et al., 1996; Kerscher et al., 1997; Davis et al., 1998; Koehler et al., 1998a; Endres et al., 1999). Tim54p and Tim22p are integral membrane proteins that associate with several proteins in the intermembrane space, including Tim12p, Tim10p, and Tim9p (Koehler et al., 1998b; Sirrenberg et al., 1998; Adam et al., 1999). Tim12p, Tim10p, and Tim9p are homologous proteins and are thought to play a role in shuttling imported proteins from the TOM complex in the OM to the Tim54p-Tim22p complex in the IM (Koehler et al., 1998a; Sirrenberg et al., 1998; Adam et al., 1999). Recently, two new members of the Tim12p-Tim10p-Tim9p family have been identified, Tim13p and Tim8p, but their role in import is not clear (Koehler et al., 1999).
To identify additional members of the Tim54p-Tim22p insertion complex, we isolated genes that, when present in multiple copies, suppress the temperature-sensitive growth defect of the tim54-1 mutant. We identified a new gene, called TIM18, which encodes an integral protein residing in the mitochondrial IM. Both genetic and biochemical experiments suggest that Tim18p is a member of the Tim54p-Tim22p complex.
MATERIALS AND METHODS
Strains and Relevant Genotypes
MATα tim54::URA3 trp1Δ63 leu2Δ1 ura3-52 strain 735 contains tim54-1 on TRP1-CEN6 plasmid pOK24 (Kerscher et al., 1997). MATa tim23-1 ura3 trp1 his3 leu2 strain 574 (Ryan et al., 1998) and MATα tim54-1 trp1Δ63 leu2Δ1 ura3-52 his3-Δ200 strain 809 (Kerscher et al., 1997) have been described. MATa/MATα trp1Δ63/trp1Δ63 his3Δ200/his3Δ200 leu2Δ1/leu2Δ1 ura3-52/ura3-52 strain 605 was constructed by crossing strain FY833 to strain FY834 (Winston et al., 1995). MATa/MATα trp1Δ63/trp1Δ63 his3Δ200/his3Δ200 leu2Δ1/leu2Δ1 ura3Δ0/ura3Δ0 strain 1082 was constructed by crossing strain BY4733 to strain BY4734 (Brachmann et al., 1998). tim54-1/tim18::HIS3 diploid strain 1009 was constructed by crossing tim54-1 strain 809 to tim18::HIS3 strain 994 (see below). Standard yeast genetic techniques and media were used (Rose et al., 1988).
Isolation of TIM18
Yeast strain 735 contains the tim54::URA3 disruption and pOK24, which carries tim54-1 on a TRP1-CEN6–containing plasmid (Kerscher et al., 1997). Strain 735 was transformed with a library containing genomic DNA fragments in the 2μ-LEU2 plasmid YEp13 (Nasmyth and Tatchell, 1980). Five thousand Leu+ transformants were selected at 24°C and then replica plated to YEP medium containing 2% glycerol and 2% ethanol as the sole carbon source. Thirty-one transformants that grew at 34°C were isolated, and the LEU2-containing plasmids were isolated. Seven of the 31 plasmids were able to rescue the tim54-1 defect upon retransformation. PCR analysis showed that four of the plasmids contained either TIM22 (three isolates) or TIM54 (one isolate). The remaining three plasmids were partially sequenced, and two plasmids contained overlapping inserts of DNA from chromosome XV. Plasmid pOK94 carries the tim54-1–suppressing activity on an ∼6-kilobase pair (kbp) genomic DNA fragment from chromosome XV. Subcloning experiments localized the tim54-1–suppressing activity to an ∼1.26-kbp EcoRI fragment containing a single ORF, YOR297c. Plasmid pOK66, which carries the YOR297 EcoRI fragment inserted into plasmid pRS423 (Sikorski and Hieter, 1989), suppressed the tim54-1 temperature-sensitive phenotype. One plasmid contained sequences from chromosome VII and has not been characterized.
Plasmid Constructions
2μ-HIS3 plasmid pOK59 carries TIM54 on a 3-kbp ClaI fragment inserted in pRS423 (Sikorski and Hieter, 1989) and was constructed by homologous recombination in yeast (Oldenburg et al., 1997) between a PvuII fragment derived from pOK31 (Kerscher et al., 1997) and XhoI-EcoRI–digested pRS423. Plasmid pJH203 (Holder, unpublished data) was constructed by insertion of an EcoRI fragment containing TIM22 from −392 to 863 into pRS423. 2μ-YLR164w plasmid pOK90 was constructed by amplifying yeast genomic DNA with the use of oligonucleotides 234 (5′-GGAATTCCCTGATTCGCACCTTC-3′) and 235 (5′-GGAATTCGTCACCACTAACCAAAC-3′) and PCR (Saiki et al., 1985). After EcoRI digestion, the PCR product was inserted into pRS426 (Sikorski and Hieter, 1989), forming pOK90. 2μ-SDH4 plasmid pOK91 was constructed by isolating a PvuI fragment containing SDH4 from plasmid SDH4-17 (a gift from B. Lemire, University of Alberta, Edmonton, Alberta, Canada) (Bullis and Lemire, 1994) and using this fragment for recombination in yeast with XhoI-BamHI–digested pRS425 (Sikorski and Hieter, 1989).
Gene Disruption of TIM18 and TIM54
A complete null mutation in the chromosomal copy of TIM18 was constructed with the use of a PCR-based gene disruption method (Lorenz et al., 1995). Briefly, a HIS3 DNA fragment was amplified with the use of oligonucleotides 221 (TCTTAGAAATGCAAA-AAAAAAGAAAAAGTATGGGTGAGTCAGATTGTACTGAGAG-TGCAC) and 222 (ATGCGAGGTGCAACAACTGAGTAATTTAATACCTTTGGTACTGTGCGGTATTTCACACCG) and plasmid pRS303 (Sikorski and Hieter, 1989) as the template. The amplified HIS3 DNA fragment, flanked by 40 base pairs (bp) of TIM18 sequences immediately upstream and downstream of the ORF, was transformed into his3/his3 diploid strains 605 and 1082. His+ diploid transformants were isolated and shown by PCR analysis to have one of two copies of TIM18 disrupted (tim18::HIS3). MATα tim18::HIS3 strain 992, MATa tim18::HIS3 strain 994, MATα TIM18 strain 993, and MATa TIM18 strain 995 are the meiotic products from tim18::HIS3/TIM18 diploid strain 987 (strain 605 background).
tim54::KAN strain 1078, which lacks the TIM54 ORF, was constructed by PCR-mediated gene replacement into diploid strain 605 with the use of oligonucleotides 382 (GCTTTAAAGTCCATTG-TTCTCAAAAGAAGCTCAATAGACCAGATTGTACTGAGTGCA-C) and 383 (CGTCGATCGTGCATGATGATAAAACATAATATATAT-CCAACTGTGCGGTATTTCACACCG) and kanMX4 plasmid pRS400 (Brachmann et al., 1998). G418-resistant transformants were then transformed with TIM54-URA3 plasmid pOK30 (Kerscher et al., 1997). After sporulation and meiotic analysis, haploid strain 1078, which carries tim54::KAN and pOK30, was isolated.
Construction of a Hemagglutinin Epitope-tagged Version of the Tim18 Protein
pOK70, which contains TIM18 with a NotI site proximal to the stop codon, was constructed as follows. An ∼1-kbp PCR product containing the TIM18 ORF was amplified from plasmid pOK66 with the use of oligonucleotides 229 (GGAATTCGTGTTAATG) and 227 (ATAGTTTAGCGGCCGCCGTTTCTTCCAAATATATAC), digested with EcoRI and NotI, and inserted into Bluescript SK II+ (Stratagene, La Jolla, CA). A 114-bp NotI fragment containing three copies of the influenza hemagglutinin (HA) epitope (Field et al., 1988; Tyers et al., 1992) was inserted into pOK70, yielding pOK73. pOK73 was digested with AatII and SacI, and an AatII-SacI fragment from pOK48 (Kerscher et al., 1997) containing the 3′-untranslated region of TIM23 was inserted to yield pOK74. TIM18-HA was isolated from pOK74 as a 2.2-kbp PvuII fragment and inserted into the HindIII-XhoI–digested LEU2-CEN6 plasmid pRS315 (Sikorski and Hieter, 1989) by homologous recombination in yeast (Oldenburg et al., 1997), forming pOK1032. pOK1032 was transformed into MATα tim18::HIS3 strain 992 (making strain 1032). 2μ-TIM18-HA plasmid pOK1030 was constructed as described for pOK1032 except that a 2.2-kbp PvuII fragment containing TIM18-HA from pOK74 was inserted into the LEU2-2μ plasmid pRS425 (Sikorski and Hieter, 1989).
Subcellular and Submitochondrial Fractionation
Yeast cells were grown to OD600 of ∼1.0 in YEP medium containing glycerol and ethanol. Cells were converted to spheroplasts and homogenized in breaking buffer (0.6 M mannitol, 20 mM HEPES/KOH, pH 7.4), and a crude cytosolic fraction and a mitochondrial pellet were isolated by centrifugation at 9600 × g for 10 min as described (Daum et al., 1982). Osmotic disruption of the mitochondrial OM was accomplished by resuspending mitochondrial pellets in 20 mM HEPES-KOH, pH 7.4, at a protein concentration of 1 mg/ml. For protease digestions, 100 μg of mitochondria at 1 mg/ml was treated with 50 μg/ml proteinase K for 20 min on ice, followed by the addition of 1 mM PMSF (Sigma Chemical, St. Louis, MO). Separation of mitochondrial OM and IM vesicles on sucrose gradients was performed as described (Emtage and Jensen, 1993). To show that Tim18p is an integral membrane protein, mitochondria were treated with either 0.1 M Na2CO3, pH 11, or 1 M NaCl and centrifuged for 30 min at ∼30 pounds per square inch in a Beckmann (Fullerton, CA) Airfuge. For analysis, proteins were separated on SDS-polyacrylamide gels (Laemmli, 1970; Haid and Suissa, 1983) and transferred (Haid and Suissa, 1983) to Immobilon P membranes (Millipore, Bedford, MA). Membranes were decorated with antibodies (Haid and Suissa, 1983), and immune complexes were detected by chemiluminescence (SuperSignal West Pico, Pierce, Rockford, IL).
Imports into Isolated Mitochondria
For in vitro transcription, TIM18 was placed behind the SP6 promoter as follows. The TIM18 ORF was amplified with the use of PCR and primers 219 (GAATTCCATGGGGAATCTGACTC) and 220 (CCGCTCGAGAGGTGCAACAACTGAG). After digestion with XhoI and EcoRI, the TIM18 PCR fragment was inserted into SalI-EcoRI–digested pSP64 (Promega, Madison, WI), forming plasmid pOK67. pOK68, which expresses a truncated version of TIM18 lacking the first 43 amino acids, TIM18(44-192), was constructed as described above except that oligonucleotide 223 (CCGCTCGAGATGGCTGAAAATAAAATAAAC) was substituted for primer 220. Labeled Tim18p and Tim18p(44–192) proteins were synthesized with the use of 1.5 mCi/ml [35S]methionine (Amersham, Arlington Heights, IL) in the SP6-Quick system (Promega) according to the manufacturer's instructions. Mitochondria for in vitro imports were isolated from strain D273-10b (Sherman, 1964) as described (Daum et al., 1982) except that SEH buffer (250 mM sucrose, 1 mM EDTA, 20 mM HEPES-KOH, pH 7.4) was sometimes used in place of breaking buffer. Fifteen microliters of reticulocyte lysate containing precursor proteins was added to 80 μg of mitochondria (1 mg/ml) in import buffer (Scherer et al., 1992) in each import reaction. Imports were stopped by incubation on ice and by the addition of valinomycin (Sigma Chemical) to 0.5 μM and carbonyl cyanide m-chlorophenylhydrazone (CCCP; Sigma Chemical) to 25 μM. To remove nonimported proteins, mitochondria were digested with 100 μg/ml trypsin (Sigma Chemical) for 30 min on ice, followed by the addition of a fivefold molar excess of soybean trypsin inhibitor (Sigma Chemical). After SDS-PAGE, labeled proteins were visualized by autoradiography (Bonner and Laskey, 1974) or exposed to a Molecular Dynamics (Sunnyvale, CA) Phosphor screen overnight, scanned with a Molecular Dynamics Storm 860 PhosphorImager, and analyzed with ImageQuant Software (Molecular Dynamics).
Immune Precipitations and Blue Native Electrophoresis
For immune precipitations, mitochondria were isolated from tim18::HIS3 strain 992 that expresses Tim18p-HA from plasmid 1032. Mitochondria were solubilized in 0.5% digitonin, 50 mM NaCl, 30 mM HEPES-KOH, pH 7.4, 1 mM PMSF, 1 mM 4-(2-aminoethyl) benzenesulfonylfluoride (Calbiochem-Novabiochem, La Jolla, CA)], 1 μg/ml leupeptin (Calbiochem-Novabiochem), and 1 μg/ml aprotinin (Sigma Chemical) at a protein concentration of 1 mg/ml. Unsolubilized material was removed from detergent lysates by centrifugation at 12,500 × g for 10 min. To 500 μl of lysate, 200 μl of a 1:1 slurry of agarose beads coupled to antibodies directed to the HA epitope was added. After rocking for 4 h at 4°C, immune complexes were recovered by centrifugation at 1300 × g for 1 min and washed. Equal amounts of pellets and supernatants were separated by SDS-PAGE and analyzed by immune blotting. HA antibodies were coupled to agarose beads with the use of the ImmunoPure IgG orientation kit (Pierce) according to the manufacturer's instructions.
For blue native gel electrophoresis, mitochondria were isolated from wild-type strain 993, tim18::HIS3 strain 992, strain 1032 that expresses Tim18p-HA, strain 494 that expresses Tim54p-HA (Kerscher et al., 1997), and strain 800 that expresses Tim22p-HA (Kerscher et al., 1997). Fifty micrograms of mitochondria from each strain was solubilized in buffer containing 1% digitonin and subjected to blue native electrophoresis as described (Schagger and von Jagow, 1991; Dekker et al., 1996, 1997).
Antiserum and Antibodies
Antiserum was raised to PiC by expressing the protein in bacteria from plasmid pNYHM131 as described (Wohlrab and Briggs, 1994). Inclusion bodies containing PiC were isolated (Harlow and Lane, 1988) and used to immunize rabbits (Covance, Denver, PA). Antibodies to the HA epitope (Niman et al., 1983) were isolated from ascites fluid prepared with the use of the 12CA5 cell line (BABCO, Berkeley, CA). Antiserum to hexokinase (Davis, unpublished data) was produced with the use of the purified protein (Sigma Chemical). Antisera to Tim54p (Kerscher et al., 1997), Tim22p (Kerscher et al., 1997), Tim23p (Emtage and Jensen, 1993), Tim17p (Ryan et al., 1998), OM45p (Yaffe et al., 1989), and Cox4p (Jensen et al., 1992) have been described.
RESULTS
TIM18 Encodes a Multiple Copy Suppressor of the tim54-1 Mutant
Part of the evidence that Tim22p interacts with Tim54p came from our previous observation that multiple copies of the TIM22 gene suppressed the temperature-sensitive growth defect of a tim54-1 mutant (Kerscher et al., 1997). To identify additional components of the Tim54p-Tim22p import complex, we screened a yeast genomic library for new high-copy suppressors of tim54-1. We transformed tim54::URA3 trp1 leu2 strain 735, which contains tim54-1 on plasmid pOK24, with a genomic library carried in the 2μ-LEU2 vector YEp13 (Nasmyth and Tatchell, 1980). Plasmids carrying the 2μ origin of replication are present in 10–40 copies per cell, resulting in overexpression of genes carried on these plasmids (Armstrong et al., 1989). Approximately 5000 Leu+ transformants were isolated at 24°C and tested for their ability to grow at 34°C. We initially identified 36 colonies that grew at 34°C, but the temperature-resistant phenotype was shown to be plasmid dependent for only 7 transformants. PCR analysis showed that three of the plasmids carried TIM22 and one of the plasmids contained TIM54. The remaining three plasmids were partially sequenced. Two plasmids contained overlapping inserts of DNA from chromosome XV, and the tim54-1–suppressing activity was localized to an ∼1.26-kbp EcoRI fragment containing a single ORF, YOR297c (Poirey et al., 1997). As described below, we found that the protein encoded by YOR297c is located in the mitochondrial IM and is part of the Tim54p-Tim22p complex. YOR297c encodes a 21.9-kDa protein that is processed to an ∼18-kDa mature form after its import into mitochondria (see below). We have named the gene TIM18 and the protein Tim18p, consistent with the nomenclature for mitochondrial import components (Pfanner et al., 1996).
To determine if the suppression activity of TIM18 is limited to tim54-1, we examined the consequence of multiple copies of TIM18 on the tim23-1 mutant. The tim23-1 mutant was transformed with 2μ plasmids containing TIM18, TIM23, TIM17, or an empty vector, and we compared the results with those of tim54-1 cells transformed with 2μ plasmids carrying TIM18, TIM54, TIM22, or an empty vector (Figure 1A). In the tim54-1 mutant, all transformants grew at 24°C, but only cells that contained TIM54, TIM22, or TIM18 grew at 34°C. Multiple copies of either TIM18 or TIM22 suppressed the growth defect of the tim54-1 mutant. In contrast, TIM18 did not allow tim23-1 cells to grow at 34°C. Only TIM17- or TIM23-containing plasmids rescued the growth defect of tim23-1. Therefore, we found that there is a specific genetic interaction between TIM18 and tim54-1 similar to that seen with TIM22. Our results raise the possibility that Tim18p is part of the Tim54p-Tim22p pathway.
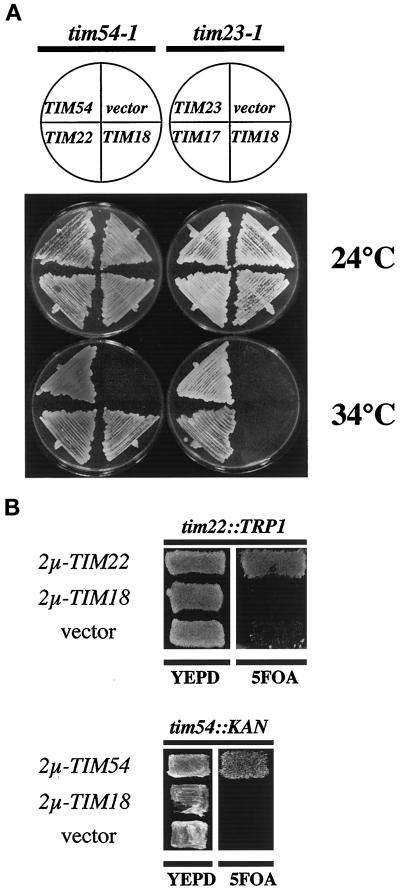
Figure 1.
(A) Multiple copies of TIM18 suppress the growth defect of tim54-1 mutants but not tim23-1 mutants. tim54-1 his3 strain 735 was transformed with one of the following high-copy plasmids: 2μ-TIM18 plasmid pOK66, 2μ-TIM54 plasmid pOK59, 2μ-TIM22 plasmid pJH203, or the empty vector pRS423 (Sikorski and Hieter, 1989). tim23-1 leu2 ura3 strain 574 was transformed with 2μ-TIM18 plasmid pOK66, 2μ-TIM23 plasmid pKR21 (Ryan et al., 1998), 2μ-TIM17 plasmid pKR7 (Ryan et al., 1994), or the empty vector pRS426 (Sikorski and Hieter, 1989). Transformants were streaked onto YEPmedium containing 2% glycerol and ethanol and incubated at 24 or 34°C for 5 d. (B) Multiple copies of TIM18 do not suppress the tim54::KAN or tim22::TRP1 disruptions. tim22::TRP1 strain 935, which carries the TIM22-URA3 plasmid pJH202 (Kerscher et al., 1997), was transformed with 2μ-TIM18 plasmid pOK66, 2μ-TIM22 plasmid pJH203, or the empty vector pRS423 (Sikorski and Hieter, 1989). tim54::KAN strain 1078, which carries the TIM54-URA3 plasmid pOK30 (Kerscher et al., 1997), was transformed with 2μ-TIM18 plasmid pOK66, CEN-TIM54 plasmid pOK22 (Kerscher et al., 1997), or the empty vector pRS424 (Sikorski and Hieter, 1989). Transformants were patched onto YEPD medium and then replica plated onto medium containing 5-FOA to select for loss of the TIM22-URA3 or TIM54-URA3 plasmids.
Although multiple copies of TIM18 allow the temperature-sensitive tim54-1 mutant to grow at 34°C, high levels of Tim18p do not completely bypass the requirement for Tim54p or Tim22p. As shown in Figure 1B, the 2μ-TIM18 plasmid did not allow strains carrying a tim54::KAN or a tim22::TRP1 disruption to grow in the absence of TIM54 or TIM22, respectively.
Tim18p Is Required for Wild-Type Cell Growth
To examine the function of the Tim18 protein, we constructed a complete disruption of TIM18 coding sequences with HIS3. We identified transformants in which one of the two copies of TIM18 was replaced by the tim18::HIS3 disruption. His+ diploid cells were sporulated, and the haploid progeny were allowed to grow at 24, 30, or 34°C on rich medium containing glucose as the carbon source (Figure 2A). When spores were grown at 34°C, all four meiotic progeny germinated and formed colonies of approximately equal size. In each tetrad, two of the colonies were His+ and were assumed to carry the tim18::HIS3 disruption, whereas the other two colonies were His− and thus carried the wild-type TIM18 gene. At 30°C, tim18::HIS3 cells formed colonies but grew slower than wild-type cells. Our results indicate that TIM18 is not an essential gene.
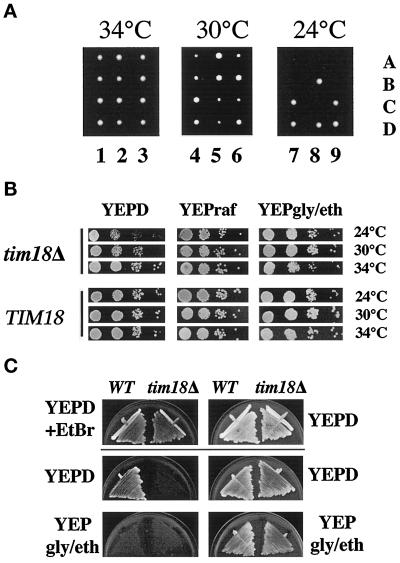
Figure 2.
TIM18 is required for normal cell growth. (A) tim18::HIS3 disruptions are cold sensitive for growth. The tim18::HIS3 disruption in diploid strain 605 was sporulated, and meiotic progeny were allowed to grow on YEP medium containing 2% glucose for 5 d at 34, 30, or 24°C. Lanes 1–9 show different tetrads, and individual spores are labeled A–D. (B) The cold-sensitive growth defect of tim18::HIS3 is limited to glucose medium. tim18::HIS3 strain 994 and TIM18 strain 993 were pregrown at 34°C on YEP medium containing 2% glucose (YEPD), YEP medium containing 2% raffinose (YEPraf), or YEP medium containing 2% glycerol and ethanol (YEPgly/eth). Two OD600 units of cells were diluted in 10-fold increments, and 10 μl of each dilution (starting with the 1:100 dilution) was spotted onto YEPD, YEPraf, or YEPgly/eth medium and incubated at 24, 30, or 34°C for 3–8 d. (C) tim18::HIS3 cells cannot tolerate loss of mitochondrial DNA. TIM18 and tim18::HIS3 cells were grown at 30°C on YEPD medium in the presence (YEPD+EtBr) or absence (YEPD) of 25 μg/ml ethidium bromide, streaked onto YEPD or YEPgly/eth medium lacking ethidium bromide, and incubated at 30°C for 5 d. WT, wild type.
We found that tim18::HIS3 mutants are cold sensitive for growth on glucose-containing medium. When tetrads from tim18::HIS3/TIM18 diploids were grown at 24°C, only two of the four spores grew into distinct colonies (Figure 2A). These colonies were shown to be His− and therefore contained the wild-type TIM18 gene. His+ spores containing tim18::HIS3 germinated, grew into very small colonies, and then arrested in their growth after about eight divisions. The microcolonies formed from these spores did not undergo additional growth even after prolonged incubation. We found that the cold-sensitive growth defect was limited to glucose-containing medium. As shown in Figure 2B, tim18::HIS3 cells pregrown at 34°C and then spotted onto glucose-containing medium displayed a clear growth defect at 24°C compared with wild-type cells. At 30°C, the growth defect of tim18::HIS3 was detectable, but it was less than at 24°C. By comparison, tim18::HIS3 cells grown on medium containing raffinose or glycerol/ethanol medium grew at rates indistinguishable from those of wild-type cells at all temperatures tested.
tim18::HIS3 disruptions in a second strain background displayed a more extreme growth defect than those in the 605 background (not shown). Haploid tim18::HIS3 cells derived from strain 1082 were inviable on glucose medium at 18, 24, 30, and 34°C. tim18::HIS3 cells were able to grow on glycerol or raffinose medium, although they grew at reduced rates compared with wild-type cells. The reason that tim18::HIS3 disruptions show a strain-specific phenotype is not known. However, disruptions of other yeast genes, such as CHC1 (Munn et al., 1991) and HRD2 (Yokota et al., 1996), often yield distinct results in different strain backgrounds.
We found that tim18::HIS3 cells cannot tolerate the loss of mitochondrial DNA (Figure 2C). tim18::HIS3 and TIM18 cells were grown for 3 d at 30°C on medium containing 25 μg/ml ethidium bromide (YEPD+EtBr) to induce loss of mitochondrial DNA (Goldring et al., 1970) and then tested for growth on different carbon sources. TIM18 cells were able to grow on glucose medium (YEPD) after ethidium bromide treatment but were unable to grow on nonfermentable medium (YEPgly/eth) as a result of loss of mitochondrial DNA. In contrast, tim18::HIS3 cells pregrown on ethidium bromide failed to grow on either YEPD or YEPgly/eth medium. We conclude that tim18::HIS3 cells that have lost mitochondrial DNA are inviable.
tim18::HIS3 Is Lethal in Combination with tim54-1
Because multiple copies of TIM18 suppress the temperature-sensitive growth defect of the tim54-1 mutant, Tim18p, like the Tim54 protein, may play a role in import. Supporting this idea, we found that tim18::HIS3 tim54-1 double mutants were inviable, exhibiting a synthetic-lethal phenotype. tim18::HIS3/tim54-1 diploid strain 1009 was sporulated, and the meiotic progeny were allowed to grow at 24°C on medium containing glycerol and ethanol (Figure 3A). In 20 tetrads (9 are shown in Figure 3A), we failed to recover any progeny carrying both tim54-1 and tim18::HIS3. Spores inferred to be double mutants germinated but arrested their growth after a few divisions. The result that the combination of tim18::HIS3 and tim54-1 is lethal provides additional genetic evidence for an interplay between Tim18p and Tim54p. tim18::HIS3 did not exhibit synthetic lethality with tim23-1 (Kerscher, unpublished observations), indicating that the interaction of Tim18p and Tim54p is specific.
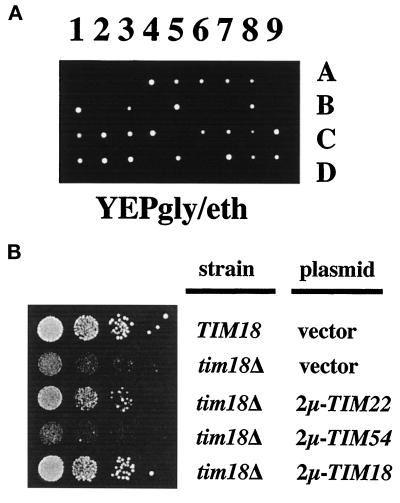
Figure 3.
TIM18 genetically interacts with both TIM54 and TIM22. (A) tim18::HIS3 is lethal in combination with tim54-1. tim54-1/tim18::HIS3 diploid strain 1009 was sporulated, and meiotic progeny were allowed to grow for 8 d at 24°C on YEP medium containing 2% glycerol and ethanol (YEPgly/eth). Lanes 1–9 show different tetrads, and individual spores are labeled A–D. (B) Multiple copies of TIM22 suppress the growth defect of tim18::HIS3. tim18::HIS3 strain 992 (tim18Δ) was transformed with the following high-copy plasmids: 2μ-TIM22 plasmid pJH202 (Kerscher et al., 1997), 2μ-TIM54 plasmid pOK31 (Kerscher et al., 1997), 2μ-TIM18 plasmid pOK94, or the empty vector pRS426. Wild-type TIM18 ura3 strain 993 was transformed with the empty vector pRS426. Cells were grown at 34°C in YEPD, diluted in 10-fold steps, spotted onto YEPD plates at 24°C, and incubated for 5 d.
Multiple Copies of TIM22 Suppress tim18::HIS3
We previously showed that the tim54-1 mutant was suppressed by high levels of TIM22 expression (Kerscher et al., 1997). We found a similar interaction between tim18::HIS3 and TIM22. We transformed tim18::HIS3 cells with 2μ-containing plasmids carrying either TIM18, TIM22, TIM54, or an empty vector and tested the cells for growth at 24°C on glucose-containing medium. As shown in Figure 3B, tim18::HIS3 cells containing either the TIM18 or TIM22 gene grew similar to wild-type cells on YEPD at 24°C. High levels of TIM54, on the other hand, did not suppress the cold-sensitive growth defect of tim18::HIS3. Our results show that the function of Tim18p can be bypassed by increased levels of Tim22p and suggest that Tim18p, like Tim22p, is part of the protein import pathway.
Tim18p Is Homologous to Two Other Yeast Proteins
The predicted protein from the TIM18 sequence (yeast ORF YOR297c) is a protein of 21.9 kDa (Figure 4A). Hydropathy analysis suggested that Tim18p is a membrane protein with three potential membrane-spanning segments (Figure 4A, underlined amino acids). In addition, the amino terminus of Tim18p has many of the characteristics of a mitochondrial presequence (Roise and Schatz, 1988; Claros and Vincens, 1996). The first 44 amino acids of Tim18p contain five basic residues, no acidic amino acids, numerous polar residues, and no long stretches of hydrophobic amino acids. When plotted on a helical wheel (Claros and Vincens, 1996), the basic residues of the amino terminus cluster to one side of the helix. Tim18p is predicted to contain a matrix processing protease site at amino acid position 44/45 (Claros and Vincens, 1996).
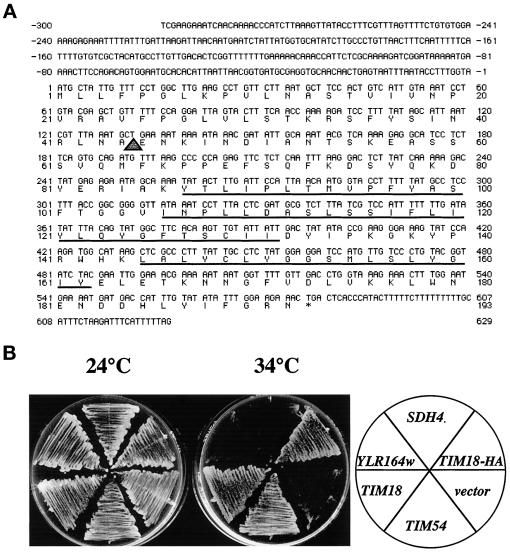
Figure 4.
The Tim18 protein is homologous to two other yeast proteins. (A) DNA sequence of TIM18 and its predicted protein product. The underlined amino acids represent potential membrane-spanning domains predicted from hydropathy analysis. The triangle indicates a potential mitochondrial processing protease cleavage site. (B) Increased levels of Sdh4p and YLR164w, proteins homologous to Tim18p, do not suppress the tim54-1 growth defect. tim54-1 strain 809 was transformed with one of the following high-copy plasmids: 2μ-SDH4 plasmid pOK91, 2μ-YLR164w plasmid pOK90, 2μ-TIM18 plasmid pOK66, 2μ-TIM18-HA plasmid pOK1030, or the empty vector pRS423. Transformants were streaked onto YEP medium containing 2% glycerol and ethanol and incubated at 24°C for 5 d or at 34°C for 3 d.
The Tim18 protein is homologous to two other proteins in the yeast genome. One of these proteins, Sdh4p, is 39.5% identical to Tim18p and has been reported to encode a membrane anchor for the mitochondrial IM succinate dehydrogenase complex (Bullis and Lemire, 1994). The other protein is 36.4% identical to Tim18p and is an uncharacterized ORF (YLR164w). Both SDH4p and Ylr164p are 52.6% identical to each other. The two Tim18p homologues, however, do not suppress the tim54-1 mutant (Figure 4B). SDH4 and YLR164w were cloned into 2μ-containing plasmids, transformed into the tim54-1 mutant, and tested for their ability to rescue the growth defect of tim54-1 at 34°C. Multiple copies of SDH4 or YLR164w did not allow the tim54-1 strain to grow at the nonpermissive temperature. Only cells that expressed high levels of Tim18p, or a Tim18p-HA fusion protein (see below), grew at 34°C.
Tim18p Is Located in the Mitochondrial IM with Its Carboxyl Terminus Facing the Intermembrane Space
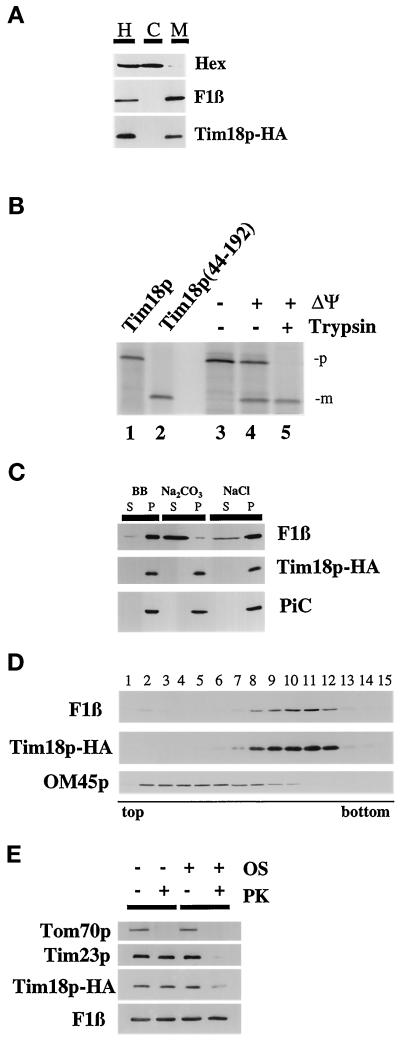
To identify the function of Tim18p, we determined the location of Tim18p in the cell. First, we inserted a HA epitope tag at the carboxyl terminus of Tim18p, forming Tim18p-HA. We found that Tim18p-HA is functional, because the growth defect of the tim18::HIS3 disruption is rescued by Tim18p-HA (Kerscher, unpublished results) and multiple copies of TIM18-HA suppress the growth defect of the tim54-1 mutant (Figure 4B). Cells expressing Tim18p-HA were homogenized and separated into a mitochondrial fraction and a postmitochondrial supernatant (Figure 5A). We found by immune blotting that Tim18p-HA cofractionated with the β-subunit of the F1-ATPase (F1β), a mitochondrial protein. No Tim18p-HA was found in the supernatant along with the cytosolic hexokinase protein.
Figure 5.
TIM18 encodes a mitochondrial inner membrane protein with its carboxyl terminus facing the mitochondrial intermembrane space. (A) Tim18p is a mitochondrial protein. tim18::HIS3 strain 992 containing plasmid pOK1032, which expresses the Tim18p-HA fusion protein, was grown in YEP medium containing 2% glycerol and ethanol. Cells were homogenized (H) and separated into a mitochondrial pellet fraction (M) and crude cytosol (C) by centrifugation. Aliquots of homogenate, mitochondria, and cytosol representing cell equivalents were subjected to SDS-PAGE. Immune blots were decorated with antibodies to the HA epitope (Tim18p-HA),antiserum to the β-subunit of the F1-ATPase (F1β), or antiserum to hexokinase (Hex) and detected by chemiluminescence. (B) Tim18p is imported into mitochondria and processed to a mature form. Mitochondria were isolated from wild-type strain D273-10b and incubated for 30 min at 30°C with the 35S-labeled Tim18 protein. After all manipulations, mitochondria were reisolated by centrifugation and analyzed by SDS-PAGE and phosphorimaging. Lane 1, 20% of the Tim18p precursor added to each import reaction. Lane 2, translation product of Tim18p(44–192), a truncated version of Tim18p lacking its first 43 amino acids. Lane 3, import in the presence of 25 μM CCCP and 0.5 μM valinomycin (−ΔΨ). Lane 4, import. Lane 5, import followed by digestion with 100 μg/ml trypsin for 30 min. The precursor (p) and mature (m) forms of Tim18p are indicated. (C) Tim18p-HA is an integral membrane protein. Tim18p-HA mitochondria were treated with either 0.1 M sodium carbonate or 1.5 M sodium chloride or left untreated (BB) and then separated into supernatant (S) and pellet (P) fractions by centrifugation. After SDS-PAGE, proteins were analyzed by immune blotting with antibodies to the HA epitope (Tim18p-HA), F1β, a peripheral membrane protein, and PiC, an integral membrane protein. (D) Tim18p-HA is located in the mitochondrial IM. Tim18p-HA mitochondria were sonicated, and membrane vesicles were loaded on sucrose step gradients. After centrifugation, fractions were collected and analyzed by immune blotting with antibodies to the HA epitope (Tim18p-HA), the IM protein F1β, or the OM protein OM45. Fraction 1 represents the top of the gradient. (E) The carboxyl terminus of Tim18p-HA faces the IMS. Tim18p-HA mitochondria were digested with 50 μg/ml proteinase K (PK) for 30 min on ice and analyzed by immune blotting with antibodies to the HA epitope (Tim18p-HA), F1β, Tim23p, or the OM protein Tom70p. To expose proteins located in the IMS, the mitochondrial OM was ruptured by osmotic shock (OS) and then treated with protease.
Supporting our localization data, we found that Tim18p was imported into isolated mitochondria. Tim18p synthesized in vitro yielded an ∼22-kDa 35S-labeled protein after SDS-PAGE (Figure 5B, lane 1). When incubated with energized mitochondria, Tim18p was processed to an ∼18-kDa protein (Figure 5B, lane 4) that was protected from externally added protease (Figure 5B, lane 5). When the potential across the mitochondrial IM was dissipated with CCCP, import and processing of Tim18p were inhibited (Figure 5B, lane 3). Tim18p is predicted to contain a mitochondrial processing protease site at amino acid position 44/45 (Claros and Vincens, 1996). To test this possibility, we constructed a truncated version of Tim18p, Tim18p(44–192), lacking its first 43 amino acids. In vitro synthesized Tim18p(44–192) comigrates with the imported and processed form of Tim18p (Figure 5B, compare lanes 2 and 5). Our results thus indicate that Tim18p is a mitochondrial protein with an amino-terminal cleavable presequence.
Tim18p is located in the mitochondrial IM. First, mitochondria were isolated from cells expressing Tim18p-HA and treated with either sodium carbonate or sodium chloride (Figure 5C). Tim18p-HA and the IM-localized PiC protein (Phelps et al., 1991) remained in the membrane pellet after carbonate or salt treatment. In contrast, F1β, a peripheral membrane protein (Chen and Douglas, 1987), was found in the supernatant after carbonate treatment. To determine in which of the two mitochondrial membranes Tim18p resides, we prepared membrane vesicles from Tim18p-HA mitochondria and separated them into OM and IM fractions on sucrose gradients. As shown in Figure 5D, Tim18p-HA cofractionates with the IM vesicle fraction, along with the F1β protein. OM45p, a marker for the OM (Yaffe et al., 1989), migrates near the top of the gradient in fractions separate from Tim18p-HA and F1β.
The carboxyl terminus of Tim18p faces the intermembrane space (IMS). Tim18p-HA mitochondria were treated with protease either in the presence or in the absence of an intact OM (Figure 5E). Immune blots showed that the IM proteins Tim18p-HA, F1β, and Tim23p were all resistant to digestion in intact mitochondria. In contrast, Tom70p, an OM protein that faces the cytosol is removed by protease treatment. When the mitochondrial OM was disrupted by osmotic shock, Tom70p, Tim23p, and Tim18p-HA were all digested, and only F1β, which faces the matrix, was protected from the protease. Our antibodies to Tim23p recognize a domain that faces the IMS, which is accessible to digestion in mitoplasts (Ryan et al., 1998). Therefore, we conclude that the carboxyl-terminal HA tag on Tim18p-HA is located in the IMS.
Tim18p Is Part of the Tim54p-Tim22p Complex in the IM
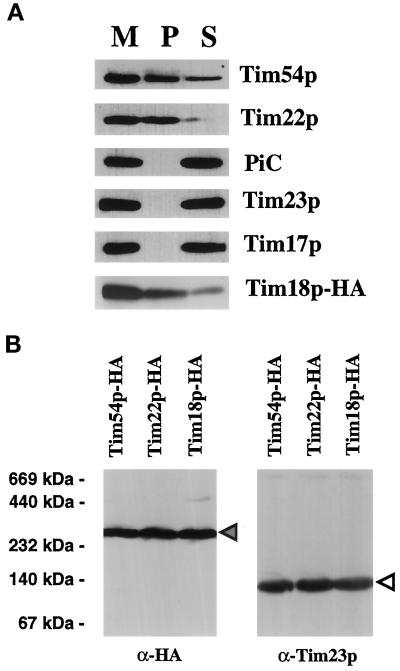
The genetic experiments described above suggest that Tim18p physically interacts with Tim54p and Tim22p. To test this idea directly, we asked if Tim54p or Tim22p could be immune precipitated along with Tim18p. Tim18p-HA mitochondria were solubilized with digitonin-containing buffer, and Tim18p-HA was precipitated with antibodies to the HA epitope (Figure 6A). Using antibodies to the HA epitope to decorate immune blots, we found that the majority of Tim18p-HA was located in the pellet. Only a small amount of the Tim18p-HA was not precipitated by the HA antibodies and remained in the supernatant. When we examined our fractions with antibodies to Tim22p, we found that virtually all of the Tim22 protein coprecipitated with Tim18-HA, indicating that Tim22p physically associates with the Tim18 protein in mitochondria. Using antibodies to Tim54p, we found that part, but not all, of Tim54p interacts with Tim18p. Only about half of Tim54p appeared to interact tightly with Tim18p. This result was similar to the results of previous experiments, in which we found that half of total Tim54p associated with Tim22p (Kerscher et al., 1997). We also found that the Tim23p or Tim17p proteins did not coprecipitate with Tim18p-HA, indicating that Tim18p's interaction with Tim22p and Tim54p is specific (Figure 6A). In addition, an abundant IM protein not involved in import, PiC, did not associate with Tim18p-HA.
Figure 6.
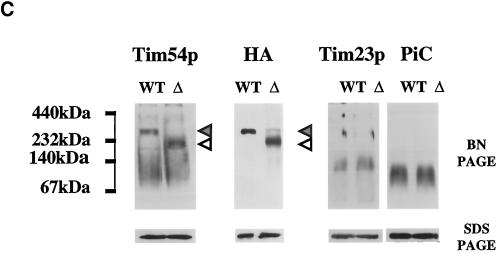
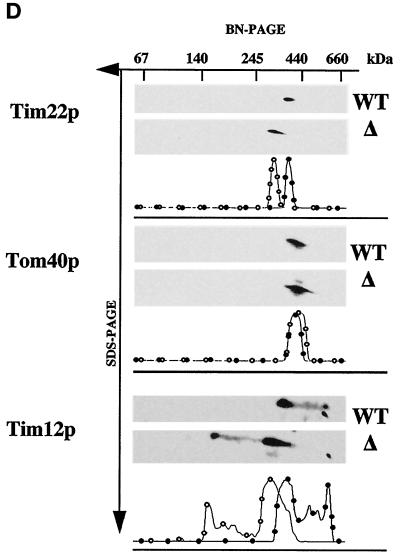
The Tim18 protein is part of the Tim22p-Tim54p complex. (A) Tim54p and Tim22p coprecipitate with Tim18p-HA. Tim18p-HA mitochondria were solubilized in 0.5% digitonin buffer. Mitochondrial extracts (M) were immune precipitated with antibodies against the HA epitope. Immunoprecipitates (P) and supernatants (S) were analyzed by SDS-PAGE, and duplicate samples were immune blotted with antibodies to the HA epitope, Tim22p, Tim54p, Tim23p, Tim17p, and PiC and then detected by chemiluminescence. (B) Tim18p-HA, Tim54p-HA, and Tim22p-HA are part of an ∼300-kDa complex. Mitochondria were isolated from cells expressing Tim18p-HA, Tim54p-HA, or Tim22p-HA, solubilized in 1% digitonin buffer, and separated by blue native electrophoresis (6–16.5% polyacrylamide). Immune blots were decorated with antibodies to HA or with antiserum to Tim23p. The protein complexes at ∼300 kDa (dark arrowhead) and ∼90 kDa (white arrowhead) are indicated. The molecular masses of standards run in adjacent lanes are indicated. (C) Tim18p is required for formation of the ∼300-kDa complex containing Tim54p. Strain 494, which expresses Tim54p-HA, TIM18 (wild-type) strain 993, tim18::HIS3 strain 992, and strain 992 carrying TIM54-HA plasmid pJH301 (Kerscher et al., 1997) were grown at 30°C on either galactose- or glycerol/ethanol-containing medium. Isolated mitochondria were solubilized in 1% digitonin buffer and separated by blue native electrophoresis (BN-PAGE). Mitochondria were also subjected to SDS-PAGE. Immune blots were decorated with antibodies to Tim54p, the HA epitope of Tim54p (HA), Tim23p, or PiC. The dark arrowheads indicate an ∼300-kDa complex in wild-type (WT) mitochondria, and the white arrowheads mark an ∼200-kDa complex in tim18::HIS3 (Δ) mitochondria. (D) tim18::HIS3 mitochondria contain an ∼200-kDa complex containing Tim22p and Tim12p. Mitochondria from TIM18 (WT) strain 993 and tim18::HIS3 (Δ) strain 992 were first subjected to blue native electrophoresis (BN-PAGE) and then to SDS-PAGE. Immune blots were decorated with antibodies to Tim22p, Tim12p, or Tom40p. Films were quantitated by scanning densitometry, and the results are shown below each pair of immune blots. (●) WT; (○) Δ.
Supporting our observations that Tim18p is a member of the Tim54p-Tim22p complex, we found that all three proteins were present in an ∼300-kDa complex after blue native electrophoresis (Figure 6B). Mitochondria isolated from cells expressing Tim18p-HA, Tim54p-HA, or Tim22p-HA were solubilized with digitonin-containing buffer, and the membrane protein complexes were separated on blue native polyacrylamide gradient gels. The mobility of the different proteins was monitored by immune blotting with antibodies to the HA epitope. Tim18p-HA comigrated with Tim54p-HA and Tim22p-HA, both of which were previously shown to be part of a 300-kDa complex (Koehler et al., 1998b; Kurz et al., 1999). Tim18p was not found in an ∼90-kDa complex containing Tim23p and Tim17p (Dekker et al., 1997). Tim54p, Tim22p, and Tim18p thus appear to be part of the same translocon.
Also supporting our conclusion that Tim18p associates with Tim54p and Tim22p, we found that the mobility of the Tim54p-Tim22p complex was altered in mitochondria lacking Tim18p (Figure 6C). Mitochondria isolated from wild-type (WT) and tim18::HIS3 (Δ) cells were subjected to blue native electrophoresis. In tim18::HIS3 mitochondria, we found that Tim54p and a Tim54p-HA fusion protein migrated in a complex of ∼200 kDa, in contrast to the ∼300-kDa complex seen with wild-type mitochondria. Other IM complexes, such as the Tim23p-Tim17p translocon or the PiC dimer, were not affected by the absence of Tim18p. To examine the distribution of Tim22p, mitochondria from wild-type or tim18::HIS3 cells were first subjected to blue native electrophoresis, and the lane was then cut out and analyzed by SDS-PAGE. Immune blots showed that Tim22p was located in an ∼300-kDa complex in wild-type mitochondria and in an ∼200-kDa complex in mitochondria lacking Tim18p. In contrast, the OM protein, Tom40p, was located in an ∼400-kDa complex (Dekker et al., 1997) in both wild-type and tim18::HIS3 mitochondria.
Several proteins of the intermembrane space, including Tim12p, Tim10p, and Tim9p, interact with the Tim54p-Tim22p complex (Koehler et al., 1998a,b; Sirrenberg et al., 1998; Adam et al., 1999). All of Tim12p is part of the TIM complex, whereas only a fraction of Tim10p and Tim9p associate with Tim54p and Tim22p. We found that the interaction of Tim12p with the TIM complex does not require Tim18p. As shown in Figure 6D, Tim12p is located in an ∼300-kDa complex containing Tim54p and Tim22p in wild-type mitochondria and in an ∼200-kDa complex in tim18::HIS3 mitochondria.
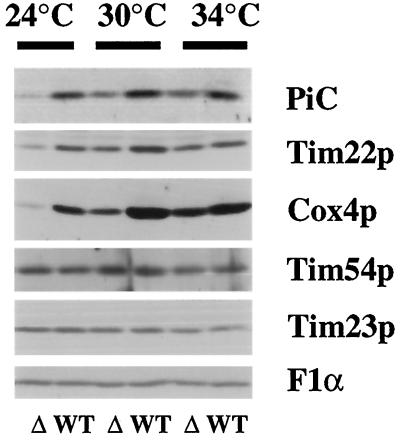
tim18::HIS3 Cells Are Cold Sensitive for the Import or Stability of Several IM Proteins
Because Tim18p is part of the Tim54p-Tim22p complex, it is possible that Tim18p also plays a role in the import of proteins into mitochondria. Tim54p and Tim22p are required for the insertion of a number of polytopic proteins into the IM, including PiC and the Tim22 protein (Sirrenberg et al., 1996; Kerscher et al., 1997). However, we found that mitochondria isolated from tim18::HIS3 cells were not defective in the import of several proteins, including PiC and Cox4p (Kerscher, unpublished observations). To determine if Tim18p plays a more subtle role in import, we took advantage of the cold-sensitive growth defect of tim18::HIS3 cells grown on glucose-containing medium. We grew wild-type and tim18::HIS3 strains in YEPD medium at 34°C, at which tim18::HIS3 cells grow at wild-type rates, and then shifted the cells to 24°C, at which tim18::HIS3 cells grow very slowly. For comparison, we also grew TIM18 and tim18::HIS3 cells at 30 and 34°C. Total protein was then isolated from these cells, and the level of different mitochondrial proteins was determined by immune blotting. As shown in Figure 7, tim18::HIS3 cells grown at 34°C contained similar amounts of all mitochondrial IM proteins examined, including PiC, Tim22, subunit IV of cytochrome oxidase (Cox4p), Tim54p, Tim23p, and the α-subunit of the F1ATPase (F1α). In contrast, tim18::HIS3 cells grown at 24°C contained greatly reduced levels of PiC, Tim22p, and Cox4p compared with wild-type cells. At 30°C, the levels of PiC, Tim22, and Cox4p in tim18::HIS3 cells were reduced, but the amounts were higher than in tim18::HIS3 cells grown at 24°C. Cells lacking Tim18p appear to be cold sensitive for the import or stability of at least several IM proteins. Whether this defect is a direct consequence of the lack of Tim18p or an indirect effect (e.g., caused by the reduced amount of Tim22p) requires further study. Our results also indicate that Tim18p may not be required for import of all IM proteins, because we found wild-type levels of Tim54p, Tim23p, and F1α in tim18::HIS3 cells grown at 24, 30, or 34°C.
Figure 7.
tim18::HIS3 cells contain reduced steady-state levels of several IM proteins. TIM18 (WT) strain 995 and tim18::HIS3 (Δ) strain 994 were grown at 34°C in YEPD medium, diluted, and then grown at 24, 30, or 34°C to an OD600 of ∼1.5. Total protein was isolated and separated by SDS-PAGE. Immune blots (75 μg of protein) were decorated with antibodies to PiC, Tim22p, Cox4p, Tim54p, Tim23p, or the α-subunit of the F1-ATPase (F1α) and detected by chemiluminescence.
DISCUSSION
We have identified a new mitochondrial IM protein named Tim18p. Several observations suggest that Tim18p, like Tim54p and Tim22p, plays a role in the import of proteins into mitochondria. For example, we identified TIM18 as a high-copy suppressor of the temperature-sensitive tim54-1 mutant. We previously showed that TIM22, when present in multiple copies, likewise rescues the growth defect of tim54-1 cells (Kerscher et al., 1997). Tim18p and Tim22p, therefore, both show a similar genetic interplay with Tim54p. We also found that the tim18::HIS3 disruption is lethal in combination with the tim54-1 mutation. Moreover, we showed that the cold-sensitive growth defect of the tim18::HIS3 disruption is suppressed by multiple copies of TIM22. Our results thus provide genetic evidence that Tim18p, Tim22p, and Tim54p interact and suggest that these three proteins function at a common step in the import pathway.
We also found evidence for physical interaction between Tim18p, Tim54p, and Tim22p. These proteins form a complex together, because all three proteins coimmune precipitated from mitochondrial extracts, and they comigrated as an ∼300-kDa complex during blue native electrophoresis. The 300-kDa complex is separate from an ∼90-kDa complex containing Tim23p and Tim17p and from the ∼400-kDa TOM complex in the OM (Dekker et al., 1997). Interestingly, the integrity of the 300-kDa complex requires the Tim18 protein. Blue native electrophoresis of mitochondria isolated from tim18::HIS3 cells showed that Tim54p and Tim22p migrate together as a smaller complex of ∼200 kDa, whereas other IM complexes, such as the Tim23p-Tim17p translocon, were not affected. Whether the 100-kDa decrease in size of the translocon represents the loss of multiple Tim18p subunits or the loss of other proteins dependent on Tim18p for assembly awaits further study. Regardless, our results indicate that Tim18p is a new member of the Tim54p-Tim22p translocon.
Tim18p is homologous to two proteins in yeast, Sdh4p (39.5% identical) and the uncharacterized ORF YLR164w (36.4% identical). Although both Sdh4p (Bullis and Lemire, 1994) and the YLR164w protein (Kerscher, unpublished data) are mitochondrial IM proteins, neither homologue appears to be part of the Tim54p-Tim22p-Tim18p complex. Multiple copies of SDH4 or YLR164w did not suppress the tim54-1 mutant. Furthermore, in preliminary experiments, we observed that the YLR164w and Sdh4 proteins do not immune precipitate along with Tim18p (Kerscher, unpublished data). Although the role of YLR164w is unknown, Sdh4p is proposed to function as the membrane anchor for other members of the succinate dehydrogenase complex (Bullis and Lemire, 1994). It is possible, therefore, that Tim18p plays an anchor function in the TIM complex. Arguing against this possibility, we found that Tim18p is not required for the interaction of Tim12p with Tim54p and Tim22p. Whether Tim10p, Tim9p, or other import proteins use Tim18p for their association with Tim54p-Tim22p awaits further studies.
TIM18 encodes a 21.9-kDa protein with a cleavable presequence. After its import into mitochondria, Tim18p was processed to an ∼18-kDa mature form. Tim18p is predicted to carry a cleavage site for the mitochondrial processing protease, MPP, between residues 44 and 45 (Claros and Vincens, 1996). Hydropathy analysis suggests that Tim18p contains three transmembrane segments, and Tim18p was located in the IM after mitochondrial fractionation. A HA tag inserted at the carboxyl terminus of Tim18p was digested when mitoplasts were treated with protease. We conclude that the mature form of Tim18p is inserted in the mitochondrial IM with three transmembrane segments, with its amino terminus facing the matrix and its carboxyl terminus exposed to the intermembrane space.
The genetic interactions between TIM18, TIM22, and TIM54 and our evidence for physical interaction suggest that Tim18p, like Tim22p and Tim54p, functions in the insertion of polytopic proteins into the IM. Consistent with such a role, we found that tim18 mutants contain lower steady-state amounts of several IM proteins. Nevertheless, Tim18p is not an essential protein and thus differs from Tim54p and Tim22p. Disruptions of TIM54 and TIM22 are inviable under all growth conditions (Sirrenberg et al., 1996; Kerscher et al., 1997), whereas tim18::HIS3 cells are cold sensitive for growth on glucose-containing medium. Several possibilities may explain why Tim18p is not essential under most growth conditions. For example, the function of Tim18p may be redundant, overlapping with the activity of another mitochondrial protein. In this case, we speculate that the second Tim18p-like activity is either absent or reduced when cells are grown on glucose, thus explaining why tim18::HIS3 mutants are defective only on glucose medium. An alternative possibility is that Tim18p is required for efficient, but not basal, levels of import. For example, the Tom6 and Tom7 proteins are nonessential subunits of the TOM complex and modulate the assembly and disassembly of the TOM complex in the mitochondrial OM, but their role in import can be detected only under special assay conditions (Alconada et al., 1995; Honlinger et al., 1996). A third possibility why the Tim18 protein is not essential is that Tim18p may be required for the import of some, but not all, proteins. For example, each import receptor in the mitochondrial OM mediates the import of a subset of proteins (Lithgow et al., 1995). Similarly, the TRAM protein of the endoplasmic reticulum translocon is required to translocate only some substrates (High et al., 1993). We are currently testing these possibilities by examining the import of many different proteins into tim18::HIS3 mitochondria.
Finally, our results also raise the possibility that Tim18p could play a role in the assembly of IM protein complexes. In particular, we found that tim18::HIS3 cells grown on glucose medium at 24°C contain reduced levels of Cox4p, in addition to low amounts of Tim22p and PiC. Although Tim22p and PiC are inserted into the IM by Tim54p-Tim22p (Sirrenberg et al., 1996; Kerscher et al., 1997), the Cox4p precursor carries an amino-terminal cleavable presequence and is imported into the mitochondrial matrix via the Tim23p-Tim17p complex (Emtage and Jensen, 1993). Cox4p, however, is a subunit of a complex in the IM, and its assembly requires chaperone activity, including that of the AAA complexes (Arlt et al., 1998). Mutants defective in chaperone activity of Yta10p and Yta12p, subunits of the m-AAA complex, have reduced levels of Cox4p similar to that seen in tim18::HIS3 cells. In addition, disruptions of YME1 (Thorsness et al., 1993; Weber et al., 1996), a subunit of the i-AAA complex, gives rise to a cold-sensitive growth defect on glucose-containing medium similar to tim18::HIS3. Furthermore, both tim18::HIS3 and yme1 mutants cannot tolerate the loss of mitochondrial DNA (Thorsness et al., 1993). We found in preliminary studies that the import, stability, or assembly of AAA complex subunits is not defective in tim18::HIS3 mutants (Kerscher, unpublished data). The interesting possibility that Tim18p itself is a chaperone or mediates an interaction with chaperones, such as the i-AAA or m-AAA complex, awaits future studies.
ACKNOWLEDGMENTS
We especially thank Bernard Lemire for SDH4-containing plasmids. We also thank Jeff Schatz for antisera to Tom70p and F1α and Mike Yaffe for antiserum to F1β. We thank Kathy Wilson, Carolyn Machamer, Alyson Aiken, Alison Davis, Scott Canna, Kara Cerveny, and Hiromi Sesaki for comments on the manuscript. This work was supported by grant R01-GM46803 from the U.S. Public Health Service to R.E.J.
REFERENCES
- Adam A, Endres M, Sirrenberg C, Lottspeich F, Neupert W, Brunner M. Tim9, a new component of the TIM22.54 translocase in mitochondria. EMBO J. 1999;18:313–319. doi: 10.1093/emboj/18.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alconada A, Kübrich M, Moczko M, Hönliger A, Pfanner N. The mitochondrial receptor complex: the small subunit Mom8b/Isp6 supports association of receptors with the general insertion pore and transfer of preproteins. Mol Cell Biol. 1995;15:6196–6205. doi: 10.1128/mcb.15.11.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison DS, Schatz G. Artificial mitochondrial presequences. Proc Natl Acad Sci USA. 1986;83:9011–9015. doi: 10.1073/pnas.83.23.9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt H, Steglich G, Perryman R, Guiard B, Neupert W, Langer T. The formation of respiratory chain complexes in mitochondria is under the proteolytic control of the m-AAA protease. EMBO J. 1998;17:4837–4847. doi: 10.1093/emboj/17.16.4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong KA, Som T, Volkert FC, Rose A, Broach JR. Propagation and expression of genes in yeast using 2-micron circle vectors. Biotechnology. 1989;13:165–192. [PubMed] [Google Scholar]
- Berthold J, Bauer MF, Schneider HC, Klaus C, Dietmeier K, Neupert W, Brunner M. The MIM complex mediates preprotein translocation across the mitochondrial inner membrane and couples it to the mt-Hsp70/ATP driving system. Cell. 1995;81:1085–1093. doi: 10.1016/s0092-8674(05)80013-3. [DOI] [PubMed] [Google Scholar]
- Bonner WM, Laskey RA. A film detection method for tritium-labeled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974;46:83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains from Saccharomyces cerevisiae 288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Bullis BL, Lemire BD. Isolation and characterization of the Saccharomyces cerevisiae SDH4 gene encoding a membrane anchor subunit of succinate dehydrogenase. J Biol Chem. 1994;269:6543–6549. [PubMed] [Google Scholar]
- Chen WJ, Douglas MG. The role of protein structure in the mitochondrial import pathway: analysis of the soluble F1-ATPase beta-subunit precursor. J Biol Chem. 1987;262:15598–15604. [PubMed] [Google Scholar]
- Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- Daum G, Böhni PC, Schatz G. Import of proteins into mitochondria: cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J Cell Biol. 1982;257:13028–13033. [PubMed] [Google Scholar]
- Davis AJ, Ryan KR, Jensen RE. Tim23p contains separate and distinct signals for targeting to mitochondria and insertion into the inner membrane. Mol Biol Cell. 1998;9:2577–2593. doi: 10.1091/mbc.9.9.2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker PJ, Martin F, Maarse AC, Bomer U, Muller H, Guiard B, Meijer M, Rassow J, Pfanner N. The Tim core complex defines the number of mitochondrial translocation contact sites and can hold arrested preproteins in the absence of matrix Hsp70-Tim44. EMBO J. 1997;16:5408–5419. doi: 10.1093/emboj/16.17.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker PJ, Muller H, Rassow J, Pfanner N. Characterization of the preprotein translocase of the outer mitochondrial membrane by blue native electrophoresis. Biol Chem. 1996;377:535–538. [PubMed] [Google Scholar]
- Dietmeier K, Hönlinger A, Bömer U, Dekker PJT, Eckerskorn C, Lottspeich F, Kübrich M, Pfanner N. Tom5 functionally links mitochondrial preprotein receptors to the general import pore. Nature. 1997;388:195–200. doi: 10.1038/40663. [DOI] [PubMed] [Google Scholar]
- Emtage JLT, Jensen RE. MAS6 encodes an essential inner membrane component of the yeast mitochondrial import pathway. J Cell Biol. 1993;122:1003–1012. doi: 10.1083/jcb.122.5.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endres M, Neupert W, Brunner M. Transport of the ADP/ATP carrier of mitochondria from the TOM complex to the TIM22.54 complex. EMBO J. 1999;18:3214–3221. doi: 10.1093/emboj/18.12.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field J, Nikawa J, Broek D, MacDonald B, Rodgers L, Wilson IA, Lerner RA, Wigler M. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol Cell Biol. 1988;8:2159–2165. doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick BS. Can Hsp70 proteins act as force-generating motors? Cell. 1995;80:11–14. doi: 10.1016/0092-8674(95)90444-1. [DOI] [PubMed] [Google Scholar]
- Goldring ES, Grossman LI, Krupnick D, Cryer DR, Marmur J. The petite mutation in yeast: loss of mitochondrial DNA during induction of petites with ethidium bromide. J Mol Biol. 1970;52:323–335. doi: 10.1016/0022-2836(70)90033-1. [DOI] [PubMed] [Google Scholar]
- Gratzer S, Lithgow T, Bauer RE, Lamping E, Paltauf F, Kohlwein SD, Haucke V, Junne T, Schatz G, Horst M. Mas37p, a novel receptor subunit for protein import into mitochondria. J Cell Biol. 1995;129:25–34. doi: 10.1083/jcb.129.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haid A, Suissa M. Immunochemical identification of membrane proteins after SDS-PAGE. Methods Enzymol. 1983;96:192–205. doi: 10.1016/s0076-6879(83)96017-2. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D, editors. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Hase T, Riezman H, Suda K, Schatz G. Import of proteins into mitochondria: nucleotide sequence of the gene for a 70-kDa protein of the yeast mitochondrial outer membrane. EMBO J. 1983;2:2169–2172. doi: 10.1002/j.1460-2075.1983.tb01718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann JM, Neupert W, Stuart RA. Insertion into the mitochondrial inner membrane of a polytopic protein, the nuclear-encoded Oxa1p. EMBO J. 1997;16:2217–2226. doi: 10.1093/emboj/16.9.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- High S, et al. Site-specific photocross-linking reveals that Sec61p and TRAM contact different regions of a membrane-inserted signal sequence. J Biol Chem. 1993;268:26745–26751. [PubMed] [Google Scholar]
- Hill K, Model K, Ryan MT, Dietmeier K, Martin F, Wagner R, Pfanner N. Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins. Nature. 1998;395:516–521. doi: 10.1038/26780. [DOI] [PubMed] [Google Scholar]
- Hines V, Brandt A, Griffiths G, Horstmann H, Brutsch H, Schatz G. Protein import into yeast mitochondria is accelerated by the outer membrane protein MAS70. EMBO J. 1990;9:3191–3200. doi: 10.1002/j.1460-2075.1990.tb07517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honlinger A, Bomer U, Alconada A, Eckerskorn C, Lottspeich F, Dietmeier K, Pfanner N. Tom7 modulates the dynamics of the mitochondrial outer membrane translocase and plays a pathway-related role in protein import. EMBO J. 1996;15:2125–2137. [PMC free article] [PubMed] [Google Scholar]
- Hönlinger A, et al. The mitochondrial receptor complex: Mom22 is essential for cell viability and directly interacts with preproteins. Mol Cell Biol. 1995;15:3382–3389. doi: 10.1128/mcb.15.6.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RE, Schmidt S, Mark RJ. Mutations in a 19-amino-acid hydrophobic region of the yeast cytochrome c1 presequence prevent sorting to the mitochondrial intermembrane space. Mol Cell Biol. 1992;12:4677–4686. doi: 10.1128/mcb.12.10.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RE, Yaffe MP. Import of proteins into yeast mitochondria: the nuclear MAS2 gene encodes a component of the processing protease that is homologous to the MAS1-encoded subunit. EMBO J. 1988;7:3863–3871. doi: 10.1002/j.1460-2075.1988.tb03272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O, Holder J, Srinivasan M, Leung RS, Jensen RE. The Tim54p-Tim22p complex mediates insertion of proteins into the mitochondrial inner membrane. J Cell Biol. 1997;139:1663–1675. doi: 10.1083/jcb.139.7.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler CM, Jarosch E, Tokatlidis K, Schmid K, Schweyen RJ, Schatz G. Import of mitochondrial carriers mediated by essential proteins of the intermembrane space. Science. 1998a;279:369–373. doi: 10.1126/science.279.5349.369. [DOI] [PubMed] [Google Scholar]
- Koehler CM, Leuenberger D, Merchant S, Renold A, Junne T, Schatz G. Human deafness dystonia syndrome is a mitochondrial disease. Proc Natl Acad Sci USA. 1999;96:2141–2146. doi: 10.1073/pnas.96.5.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler CM, Merchant S, Oppliger W, Schmid K, Jarosch E, Dolfini L, Junne T, Schatz G, Tokatlidis K. Tim9p, an essential partner subunit of Tim10p for the import of mitochondrial carrier proteins. EMBO J. 1998b;17:6477–6486. doi: 10.1093/emboj/17.22.6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz M, Martin H, Rassow J, Pfanner N, Ryan MT. Biogenesis of Tim proteins of the mitochondrial carrier import pathway: differential targeting mechanisms and crossing over with the main import pathway. Mol Biol Cell. 1999;10:2461–2474. doi: 10.1091/mbc.10.7.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lithgow T, Glick BS, Schatz G. The protein import receptor of mitochondria. Trends Biochem Sci. 1995;20:98–101. doi: 10.1016/s0968-0004(00)88972-0. [DOI] [PubMed] [Google Scholar]
- Lithgow T, Junne T, Wachter C, Schatz G. Yeast mitochondria lacking the two import receptors Mas20p and Mas70p can efficiently and specifically import precursor proteins. J Biol Chem. 1994;269:15325–15330. [PubMed] [Google Scholar]
- Lohret TA, Jensen RE, Kinnally KW. Tim23, a protein import component of the mitochondrial inner membrane, is required for normal activity of the multiple conductance channel, MCC. J Cell Biol. 1997;137:377–386. doi: 10.1083/jcb.137.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz MC, Muir RS, Lim E, McElver J, Weber SC, Heitman J. Gene disruption with PCR products in Saccharomyces cerevisiae. Gene. 1995;158:113–117. doi: 10.1016/0378-1119(95)00144-u. [DOI] [PubMed] [Google Scholar]
- Maarse AC, Blom J, Keil P, Pfanner N, Meijer M. Identification of the essential yeast protein MIM17, an integral mitochondrial inner membrane protein involved in protein import. FEBS Lett. 1994;349:215–221. doi: 10.1016/0014-5793(94)00669-5. [DOI] [PubMed] [Google Scholar]
- Mahlke K, Pfanner N, Martin J, Horwich AL, Hartl F-U, Neupert W. Sorting pathways of mitochondrial inner membrane proteins. Eur J Biochem. 1990;192:551–555. doi: 10.1111/j.1432-1033.1990.tb19260.x. [DOI] [PubMed] [Google Scholar]
- McAda PC, Douglas MG. A neutral metalloendoprotease involved in the processing of an F1-ATPase subunit precursor in mitochondria. J Biol Chem. 1982;257:3177–3182. [PubMed] [Google Scholar]
- Miller BR, Cumsky MG. An unusual import pathway for the precursor to yeast cytochrome oxidase subunit Va. J Cell Biol. 1991;112:833–841. doi: 10.1083/jcb.112.5.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BR, Cumsky MG. Intramitochondrial sorting of the precursor to yeast cytochrome c oxidase subunit Va. J Cell Biol. 1993;121:1021–1029. doi: 10.1083/jcb.121.5.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moczko M, Bomer U, Kubrich M, Zufall N, Honlinger A, Pfanner N. The intermembrane space domain of mitochondrial Tom22 functions as a trans binding site for preproteins with N-terminal targeting sequences. Mol Cell Biol. 1997;17:6574–6584. doi: 10.1128/mcb.17.11.6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moczko M, Gartner F, Pfanner N. The protein import receptor MOM19 of yeast mitochondria. FEBS Lett. 1993;326:251–254. doi: 10.1016/0014-5793(93)81801-6. [DOI] [PubMed] [Google Scholar]
- Moro F, Sirrenberg C, Schneider HC, Neupert W, Brunner M. The TIM17.23 preprotein translocase of mitochondria: composition and function in protein transport into the matrix. EMBO J. 1999;18:3667–3675. doi: 10.1093/emboj/18.13.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn AL, Silveira L, Elgort M, Payne GS. Viability of clathrin heavy-chain-deficient Saccharomyces cerevisiae is compromised by mutations at numerous loci: implications for the suppression hypothesis. Mol Cell Biol. 1991;11:3868–3878. doi: 10.1128/mcb.11.8.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth KA, Tatchell K. The structure of transposable yeast mating type loci. Cell. 1980;19:753–764. doi: 10.1016/s0092-8674(80)80051-1. [DOI] [PubMed] [Google Scholar]
- Niman HL, Houghten RA, Walker LE, Reisfeld RA, Wilson IA, Hogle JM, Lerner RA. Generation of protein-reactive antibodies by short peptides is an event of high frequency: implications for the structural basis of immune recognition. Proc Natl Acad Sci USA. 1983;80:4949–4953. doi: 10.1073/pnas.80.16.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg KR, Vo KT, Michaelis S, Paddon C. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 1997;25:451–452. doi: 10.1093/nar/25.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanner N. Mitochondrial import: crossing the aqueous intermembrane space. Curr Biol. 1998;8:R262–R265. doi: 10.1016/s0960-9822(98)70168-x. [DOI] [PubMed] [Google Scholar]
- Pfanner N, Craig EA, Meijer M. The protein import machinery of the mitochondrial inner membrane. Trends Biochem Sci. 1994;19:368–372. doi: 10.1016/0968-0004(94)90113-9. [DOI] [PubMed] [Google Scholar]
- Pfanner N, Douglas MG, Endo T, Hoogenraad NJ, Jensen RE, Meijer M, Neupert W, Schatz G, Schmitz UK, Shore G. Uniform nomenclature for the protein transport machinery of the mitochondrial membranes. Trends Biochem Sci. 1996;21:51–52. [PubMed] [Google Scholar]
- Phelps A, Schobert CT, Wohlrab H. Cloning and characterization of the mitochondrial phosphate transport protein gene from the yeast Saccharomyces cerevisiae. Biochemistry. 1991;30:248–252. doi: 10.1021/bi00215a035. [DOI] [PubMed] [Google Scholar]
- Poirey R, Cziepluch C, Tobiasch E, Pujol A, Kordes E, Jauniaux JC. Sequence and analysis of a 36.2 kb fragment from the right arm of yeast chromosome XV reveals 19 open reading frames including SNF2 (5′ end), CPA1, SLY41, a putative transport ATPase, a putative ribosomal protein and an SNF2 homologue. Yeast. 1997;13:479–482. doi: 10.1002/(SICI)1097-0061(199704)13:5<479::AID-YEA104>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Pollock RA, Hartl F-U, Cheng MY, Ostermann J, Horwich A, Neupert W. The processing protease of yeast mitochondria: the two cooperating components MPP and PEP are structurally related. EMBO J. 1988;7:3493–3500. doi: 10.1002/j.1460-2075.1988.tb03225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage L, Junne T, Hahne K, Lithgow T, Schatz G. Functional cooperation of mitochondrial protein import receptors in yeast. EMBO J. 1993;12:4115–4123. doi: 10.1002/j.1460-2075.1993.tb06095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassow J, Dekker PJT, van Wilpe S, Meijer M, Soll J. The preprotein translocase of the mitochondrial inner membrane: function and evolution. J Mol Biol. 1999;286:105–120. doi: 10.1006/jmbi.1998.2455. [DOI] [PubMed] [Google Scholar]
- Rassow J, Maarse AC, Krainer E, Kubrich M, Muller H, Meijer M, Craig EA, Pfanner N. Mitochondrial protein import: biochemical and genetic evidence for interaction of matrix Hsp70 and the inner membrane protein Mim44. J Cell Biol. 1994;127:1547–1556. doi: 10.1083/jcb.127.6.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roise D. Interaction of a synthetic mitochondrial presequence with isolated yeast mitochondria: mechanism of binding and kinetics of import. Proc Natl Acad Sci USA. 1992;89:608–612. doi: 10.1073/pnas.89.2.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roise D, Horvath SJ, Tomich JM, Richards JH, Schatz G. A chemically synthesized presequence of an imported mitochondrial protein can form an amphipathic helix and perturb natural and artificial phospholipid bilayers. EMBO J. 1986;5:1327–1334. doi: 10.1002/j.1460-2075.1986.tb04363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roise D, Schatz G. Mitochondrial presequences. J Biol Chem. 1988;263:4509–4511. [PubMed] [Google Scholar]
- Roise D, Theiler F, Horvath SJ, Tomich JM, Richards JH, Allison DS, Schatz G. Amphiphilicity is essential for mitochondrial presequence function. EMBO J. 1988;7:649–653. doi: 10.1002/j.1460-2075.1988.tb02859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Cold Spring Harbor. NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Ryan KR, Jensen RE. Protein translocation across mitochondrial membranes: what a long, strange trip it is. Cell. 1995;83:517–519. doi: 10.1016/0092-8674(95)90089-6. [DOI] [PubMed] [Google Scholar]
- Ryan KR, Leung RS, Jensen RE. Characterization of the inner membrane translocase complex: the Tim23p hydrophobic domain interacts with Tim17p but not with other Tim23p molecules. Mol Cell Biol. 1998;18:178–187. doi: 10.1128/mcb.18.1.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan KR, Menold MM, Garrett S, Jensen RE. SMS1, a high-copy suppressor of the yeast mas6 mutant, encodes an essential inner membrane protein required for mitochondrial protein import. Mol Biol Cell. 1994;5:529–538. doi: 10.1091/mbc.5.5.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki RK, Scharf S, Faloona F, Mullis KB, Horn GT, Erlich HA, Arnheim N. Enzymatic amplification of β-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Schagger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- Scherer PE, Manning-Krieg UC, Jenö P, Schatz G, Horst M. Identification of a 45-kDa protein at the protein import site of the yeast mitochondrial inner membrane. Proc Natl Acad Sci USA. 1992;89:11930–11934. doi: 10.1073/pnas.89.24.11930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Mutants of yeast deficient in cytochrome c. Genetics. 1964;49:39–48. doi: 10.1093/genetics/49.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R, Hieter P. A system of shuttle vectors and host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–28. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirrenberg C, Bauer MF, Guiard B, Neupert W, Brunner M. Import of carrier proteins into the mitochondrial inner membrane mediated by Tim22. Nature. 1996;384:582–585. doi: 10.1038/384582a0. [DOI] [PubMed] [Google Scholar]
- Sirrenberg C, Endres M, Folsch H, Stuart RA, Neupert W, Brunner M. Carrier protein import into mitochondria mediated by the intermembrane proteins Tim10/Mrs11 and Tim12/Mrs5. Nature. 1998;391:912–915. doi: 10.1038/36136. [DOI] [PubMed] [Google Scholar]
- Steger HF, Sollner T, Kiebler M, Dietmeier KA, Pfaller R, Trulzsch KS, Tropschug M, Neupert W, Pfanner N. Import of ADP/ATP carrier into mitochondria: two receptors act in parallel. J Cell Biol. 1990;111:2353–2363. doi: 10.1083/jcb.111.6.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart RA, Gruhler A, van der Klei I, Guiard B, Koll H, Neupert W. The requirement for matrix ATP for the import of precursor proteins into the mitochondrial matrix and intermembrane space. Eur J Biochem. 1994;220:9–18. doi: 10.1111/j.1432-1033.1994.tb18593.x. [DOI] [PubMed] [Google Scholar]
- Stuart RA, Neupert W. Topogenesis of inner membrane proteins of mitochondria. Trends Biochem Sci. 1996;21:261–267. [PubMed] [Google Scholar]
- Thorsness PE, White KH, Fox TD. Inactivation of YME1, a member of the ftsH-SEC18-PAS1-CDC48 family of putative ATPase-encoding genes, causes increased escape of DNA from mitochondria in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:5418–5426. doi: 10.1128/mcb.13.9.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M, Tokiwa G, Nash R, Futcher B. The Cln3-Cdc28 kinase complex of S. cerevisiae is regulated by proteolysis and phosphorylation. EMBO J. 1992;11:1773–1784. doi: 10.1002/j.1460-2075.1992.tb05229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber ER, Hanekamp T, Thorsness PE. Biochemical and functional analysis of the YME1 gene product, an ATP- and zinc-dependent mitochondrial protease from S. cerevisiae. Mol Biol Cell. 1996;7:307–317. doi: 10.1091/mbc.7.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F, Dollard C, Ricupero-Hovasse SL. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- Witte C, Jensen RE, Yaffe MP, Schatz G. MAS1, a gene essential for yeast mitochondrial assembly, encodes a subunit of the mitochondrial processing protease. EMBO J. 1988;7:1439–1447. doi: 10.1002/j.1460-2075.1988.tb02961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlrab H, Briggs C. Yeast mitochondrial phosphate transport protein expressed in Escherichia coli: site-directed mutations at threonine-43 and at a similar location in the second tandem repeat (isoleucine-141) Biochemistry. 1994;33:9371–9375. doi: 10.1021/bi00198a001. [DOI] [PubMed] [Google Scholar]
- Yaffe MP, Jensen RE, Guido EC. The major 45-kDa protein of the yeast mitochondrial outer membrane is not essential for cell growth or mitochondrial function. J Biol Chem. 1989;264:21091–21096. [PubMed] [Google Scholar]
- Yaffe MP, Ohta S, Schatz G. A yeast mutant temperature-sensitive for mitochondrial assembly is deficient in a mitochondrial protease activity that cleaves imported precursor polypeptides. EMBO J. 1985;4:2069–2074. doi: 10.1002/j.1460-2075.1985.tb03893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Jensen RE, Yaffe MP, Oppliger W, Schatz G. Import of proteins into yeast mitochondria: the purified matrix processing protease contains two subunits which are encoded by the nuclear MAS1 and MAS2 genes. EMBO J. 1988;7:3857–3862. doi: 10.1002/j.1460-2075.1988.tb03271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota K, et al. cDNA cloning of p112, the largest regulatory subunit of the human 26s proteasome, and functional analysis of its yeast homologue, sen3p. Mol Biol Cell. 1996;7:853–870. doi: 10.1091/mbc.7.6.853. [DOI] [PMC free article] [PubMed] [Google Scholar]