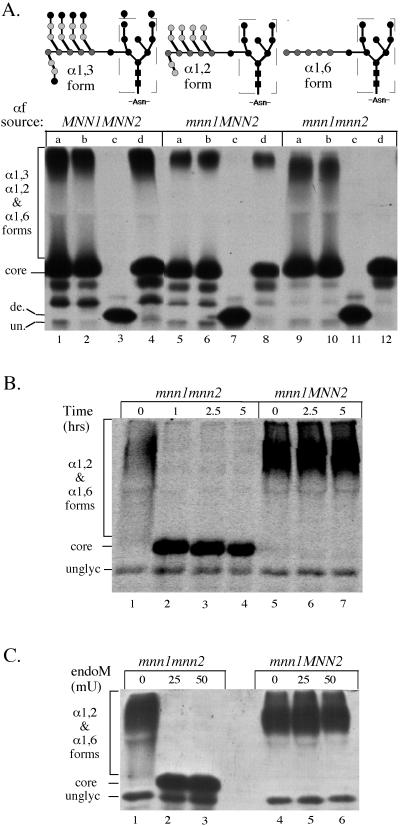
Figure 3.
Activity and specificity of endoM preparations against immunoprecipitated pro-αf. (A) MNN1 MNN2 (TBY130), mnn1 MNN2 (TBY131), and mnn1 mnn2 (TBY132) strains were labeled with 35S-amino acids for 10 min at 20°C and no chase. N-linked oligosaccharides produced from these yeast strains are represented above each genotype. Pro-αf was recovered from the cell lysates by immunoprecipition and was split into four equal samples. One sample was left untreated (lanes marked a), and the other samples were treated with the following preparations: (b) endoM buffer (mock), (c) ammonium sulfate-purified endoM (50mU), and (d) DEAE-purified endoM (50 mU). All subsequent enzyme incubations are with DEAE-purified endoM. (B and C) Pro-αf was immunoprecipitated from mnn1 and mnn1 mnn2 yeast strains labeled at 20°C for 10 min and chased for 7 min. (B) Samples were split and treated with 50 mU of endoM for the indicated times. (C)Samples were split and treated with the indicated amount of endoM. de., deglycosylated pro-αf; un., unglycosylated pro-αf. Note that the contaminating endoglycosidase apparent in lanes marked C produces a deglycosylated pro-αf with a slightly slower mobility than the unglycosylated form. This suggests cleavage between the N-acetylglucosamines leaving behind one sugar per N-linked oligosaccharide. Core indicates the ER pro-αf form carrying the oligosaccharide shown within the dashed box. All three experiments were electorphoresed on 15% SDS-polyacrylamide gels with B and C run for shorter times than A.