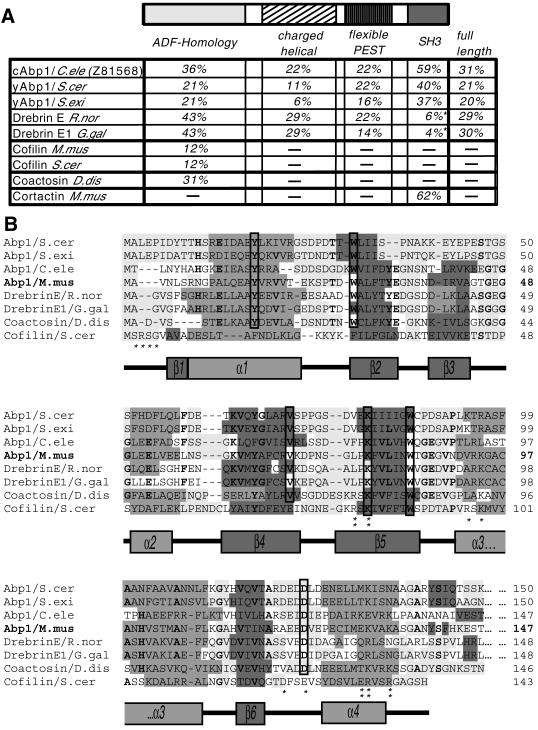
Figure 1.
Analysis of the mAbp1 sequence. (A) Domain structure of mAbp1/SH3P7 (GenBank accession number U58884) and comparison of the sequence identities of individual domains and the full-length protein of mAbp1 to corresponding areas of related proteins of the Abp1/drebrin class of ADF-H domain proteins and cortactin. Note that drebrins, in contrast to what we reported earlier (Lappalainen et al., 1998), seem not to contain a C-terminal SH3 domain. Accession numbers: S. cerevisiae Abp1 (European Molecular Biology Laboratory, X51780; Swiss-Prot, P15891), Saccharomyces exiguus Abp1 (Swiss-Prot, P38479), Rattus norvegicus drebrin (Swiss-Prot, Q07266), Gallus gallus drebrinA, E1 and E2 (Swiss-Prot, P18302), Mus musculus cofilin1 (nonmuscle isoform) (Swiss-Prot, P18760), S. cerevisiae cofilin (Swiss-Prot, Q03048), C. elegans Abp1 (ORF) (GenBank, Z81568), Dictyostelium discoideum coactosin (Swiss-Prot, P34121), M. musculus cortactin (Swiss-Prot, Q60598). The three most similar proteins in each column are highlighted in bold. (B) Identification of a common fold among Abp1s, drebrins, coactosin, and cofilin by aligning their N termini and predicting their secondary structures (light gray, flexible regions; medium gray, α-helices; dark gray, β-sheets; no underlying shading, no prediction). Predictions were performed using the PHDsec program (Rost and Sander, 1994). Residues shown by mutational analysis of yeast cofilin to be important for G and F-actin-binding (Lappalainen et al., 1997) are marked by stars; those important for F-actin binding only are marked by double stars. Residues that are >70% conserved within the ADF-H domains of Abp1s, drebrins, and coactosin are in bold, whereas residues fully conserved are boxed.