Figure 1.
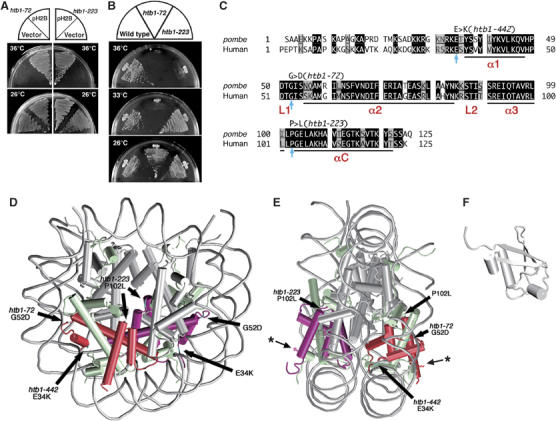
S. pombe H2B mutations reside in the conserved, non-helical regions. (A) Plasmid pH2B suppresses the ts phenotype of htb1-72 and htb1-223. (B) htb1-72 and htb1-223 mutants fail to produce colonies at 36°C on the complete YPD plates. (C) Amino-acid sequences of fission yeast and human histone H2B. Identical and similar amino acids are indicated in black and gray boxes, respectively. Substituted amino acids in the three htb1 mutants are indicated by arrows (see text). The α-helices are labeled as α1, α2, α3 and αC, whereas L1 and L2 represent the DNA binding loops. (D, E) The nucleosome structure (D, front view; E, side view; White et al, 2001) is depicted with the positions of altered amino acids. The atomic coordinates file (ID; 1ID3) was obtained from Brookhaven Protein Databank (PDB) and visualized with VMD (version 1.8.3). The DNA phosphodiester backbone is shown in gray. The octamer histone chains are schematized by the helical rods and the interhelical loops. Histone H2B subunits are indicated in red. The asterisks indicate the position of K that is associated with monoubiquitin. (F) The 3D structure of ubiquitin (PDB ID; 1UBQ) (Vijay-Kumar et al, 1987).
