Figure 2.
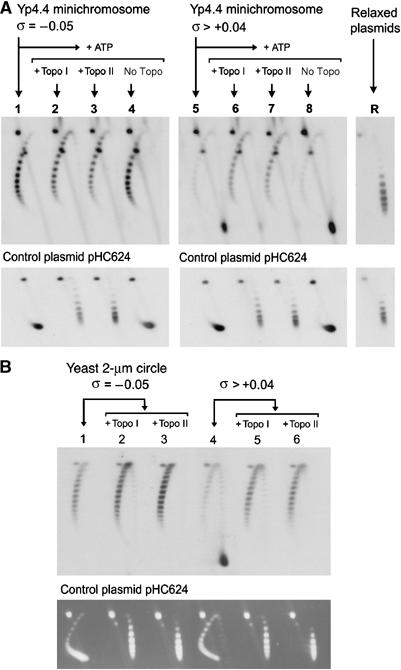
Relaxation of yeast minichromosomes by DNA topoisomerases. (A) Yeast minichromosomes, for which σ∼−0.05 (lane 1) and for which σ∼>+0.04 (lane 5) were incubated at 30°C for 30 min with catalytic amounts of vaccinia virus topo I (lanes 2 and 6), S. cerevisiae topo II (lanes 3 and 7) or no enzyme (7 and 8). Plasmids Yp4.4 and pHC624 were also relaxed in vitro (lane R). Two-dimensional electrophoresis of DNA was carried out at 25°C in a 0.8% agarose gel in TBE buffer plus 0.6 μg/ml of chloroquine at 50 V for 14 h in the first dimension (top to bottom), and TBE buffer plus 3 μg/ml of chloroquine, at 60 V for 8 h in the second dimension (left to right). Gel-blots were probed for Yp4.4 (upper panel) and for the control plasmid pHC624 (lower panel). (B) Yeast minichromosomes, for which σ∼−0.05 (lane 1) and for which σ>+0.04 (lane 4) were supplemented with an excess of control plasmid pHC624 (1 mg/ml), and incubated with catalytic amounts of S. cerevisiae topo I (lanes 2 and 5) or S. cerevisiae topo II (lanes 3 and 6) at 30°C for 30 min. After two-dimensional electrophoresis of DNA, gels were blotted and probed for the yeast 2-μm circle (upper panel) or ethidium stained to visualize pHC624 (lower panel).
