Figure 4.
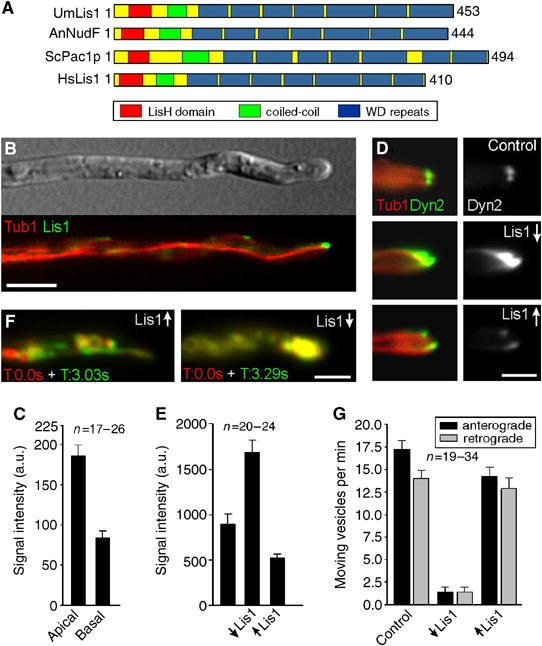
The role of a LIS1-homologue in dynein localization and endosome motility. (A) Lis1 from U. maydis (UmLis1, Accession Number: EAK84394) shares a similar domain structure with other LIS1-homologues, including those from A. nidulans (AnNudF, EAA57983), S. cerevisiae (ScPac1p, AAB00685) and human (HsLis1, NP 000421). (B) GFP-tagged endogenous Lis1 (green) localizes to plus-ends of RFP-α-tubulin containing MTs (red). Note that the tip contains the strongest signal. Bar: 5 μm. (C) Quantification of the GFP-Lis1 intensity at MT plus-ends in the apical 3 μm of the hyphal tip (apical) and in at least 12 μm distance from the tip (basal) demonstrates that Lis1 is concentrated at MT plus-ends in the tip. (D) The absence of Lis1 (Lis1↓) results in an increase of dynein at plus-ends within the hyphal tip. Overexpression of Lis1 (Lis1↑) reduces the apical dynein signal. Bar: 2 μm. (E) Quantitative analysis of GFP3Dyn2 intensity at MT plus-ends revealed that the absence of Lis1 (↓Lis1) significantly increases the amount of dynein at MT plus-ends. Consistently, Lis1 overexpression (↑Lis1) has the opposite effect. (F) Overlay of two images at different time points illustrates the motility of RFP2-tagged endosomes. Moving organelles appear in green or red, while stationary signals result in yellow color. Depletion of Lis1 led to an immobile accumulation of EE in the hyphal apex (strain FB2G3Dyn2_nLis1_YR2). Bar: 5 μm. Supplementary movies for Panel D and F are given on the EMBO web site. (G) A quantitative analysis of EE traffic reveals that overexpression of Lis1 (↑Lis1) affects EE motility only slightly, while nearly all motility is abolished in the absence of Lis1 (↓Lis1).
