Abstract
Gatifloxacin (GAT) is a new 8-methoxy fluoroquinolone with enhanced activity against gram-positive cocci. Its activity was studied in an in vitro pharmacokinetic-pharmacodynamic model against five Staphylococcus aureus strains, either susceptible to ciprofloxacin or exhibiting various levels and mechanisms of ciprofloxacin (CIP) resistance: the ATCC 25923 reference strain (MICs of CIP and GAT: 0.5 and 0.1 μg/ml, respectively), its efflux mutant SA-1 (16 and 0.5 μg/ml; mutation in the norA promoter region), and three clinical strains, Sa2102 (2 and 0.2 μg/ml), Sa2667 (4 and 0.5 μg/ml), and Sa2669 (16 and 1 μg/ml), carrying mutations in the grlA (Ser80Tyr or Phe) and gyrA (Ser84Ala) quinolone resistance-determining regions (QRDRs) for Sa2669. Plasmatic pharmacokinetic profiles after daily 1-h perfusion of 400 mg for 48 h were accurately simulated. Thus, mean maximum concentration of drug in serum values for the two administration intervals were 5.36 and 5.80 μg/ml, respectively, and the corresponding half-life at β-phase values were 8.68 and 7.80 h (goodness of fit coefficient, >0.98). Therapeutic concentrations of GAT allowed the complete eradication of the susceptible strain within 12 h (difference between the bacterial counts at the beginning of the treatment and at a defined time: −2.18 at the 1-h time point [t1] and −6.80 at t24 and t48; the bacterial killing and regrowth curve from 0 to 48 h was 30.2 h × log CFU/milliliter). However, mutants (M) with GAT MICs increased by 4- to 40-fold were selected from the other strains. They acquired mutations either supplementary (MSa2102 and MSa2667) or different (Ala84Val for MSa2669) in gyrA or in both gyrA and grlA QRDRs (MSA-1). MSa2667 additionally overproduced efflux system(s) without norA promoter modification. Thus, GAT properties should allow the total elimination of ciprofloxacin-susceptible S. aureus, but resistant mutants might emerge from strains showing reduced susceptibility to older fluoroquinolones independently of the first-step mutation(s).
Staphylococcus aureus is a major cause of community- and hospital-acquired infections. This organism is moderately susceptible to fluoroquinolones, and the extensive use of the earlier molecules of this class has often generated resistant mutants. Fluoroquinolone resistance mechanisms in S. aureus involve target alterations and overexpression of intrinsic multidrug resistance (MDR) efflux pumps (16). Indeed, quinolones exert their bactericidal activity by interacting with topoisomerases of type II, which are homologous tetramers consisting of two types of subunits, GyrA and GyrB in DNA gyrase and GrlA and GrlB in topoisomerase IV. Mutations associated with quinolone resistance primarily occur within specific sequences, termed quinolone resistance-determining regions (QRDRs), of gyrA or, preferentially in S. aureus, grlA (9, 20). Additional modifications in other topoisomerase subunits and/or in MDR systems, such as NorA, lead to increased resistance levels (22).
Gatifloxacin is a new 6-fluoro-8-methoxyquinolone that is two- to fourfold more active than ciprofloxacin against S. aureus (14, 32). Thus, this compound might become a first-line treatment of staphylococcal infections due to ciprofloxacin-susceptible strains, and it might also be useful against intermediately ciprofloxacin-susceptible strains or strains with low-level resistance which appear gatifloxacin susceptible by laboratory-based criteria. However, these in vitro data should be validated in vivo by clinical trials, including full bacteriological documentation and a statistically significant number of cases due to these particular strains. Alternatively, in vitro pharmacokinetic-pharmacodynamic (PK-PD) models might help to predict gatifloxacin efficiency in these specific instances (1, 5, 15).
The aim of this study was to use a PK-PD model previously developed in our laboratory (2) to investigate gatifloxacin efficiency on S. aureus strains either susceptible to ciprofloxacin or exhibiting various levels and genetically defined mechanisms of ciprofloxacin resistance and to determine the risk of emergence of resistant mutants.
MATERIALS AND METHODS
PK-PD model and simulated dose regimen, antibiotics, and reagents.
The two-compartment PK-PD model with full computer-controlled devices has been previously described in detail elsewhere (2). In this study, a human plasma pharmacokinetic profile of gatifloxacin was simulated after a 1-h-daily infusion of 400 mg for 48 h. Flow rates were adjusted according to reference pharmacokinetic parameters, i.e., maximum concentration of drug in serum (Cmax), 5.47 μg/ml, and half-life at β-phase (t1/2β), 7.66 h (24). All experiments were performed in cation-adjusted Mueller-Hinton broth (MH; A.D.L., Tresses, France) with concentrations of Ca2+ and Mg2+ of 23 μg/ml and 13 μg/ml, respectively, at 37°C. The antibiotic reference powders were kindly provided by their manufacturers (gatifloxacin, Grünenthal; ciprofloxacin and moxifloxacin, Bayer Pharma; norfloxacin, Merck Sharp and Dohme-Chibret). Ethidium bromide (EtBr) and reserpine were purchased from Sigma-Aldrich (Saint Quentin Fallavier, France).
Bacterial strains, antimicrobial susceptibility testing, and molecular analysis.
The reference strain of S. aureus ATCC 25923 was used to validate the model and to assess gatifloxacin pharmacodynamic parameters. SA-1 was selected from S. aureus ATCC 25923 on an EtBr concentration gradient. Three clinical strains of methicillin-susceptible S. aureus (MSSA) (Sa2667) or methicillin-resistant S. aureus (MRSA) (Sa2102 and Sa2669), isolated from the community (Sa2102) or hospitalized patients (Sa2667 and Sa2669) and exhibiting various levels of ciproflacin resistance, were also included (Table 1). MICs of gatifloxacin, ciprofloxacin, norfloxacin, moxifloxacin, and EtBr, alone and in presence of reserpine at the final concentration of 128 μg/ml, were determined in at least three independent experiments by an agar dilution method on MH medium (http://www.sfm.asso.fr). Quinolone susceptibility and resistance were defined according to the guidelines of the Antibiogram Committee of the French Society for Microbiology (e.g., ciprofloxacin MIC breakpoints for susceptible [≤1 μg/ml] and resistant [>2 μg/ml] strains) (http://www.sfm.asso.fr). After DNA chromosomal extraction, PCR amplification of the topoisomerase QRDRs and of the norA promoter region (flqB) were performed using specific pairs of primers (Table 2) and the Gold Star Taq polymerase (Applied Biosystems Division, Perkin-Elmer, Courtaboeuf, France) under standard conditions with an annealing temperature of 50°C. PCR products were then purified using microcolumns of the Microspin Sephacryl S-400 purification system (Amersham Biosciences, Buckinghamshire, England) and sequenced on both strands with an automated fluorescent method based on the dye terminator chemistry (Applied Biosystems Division, Perkin Elmer).
TABLE 1.
Characteristics of the parental and derived mutant strains of S. aureus included in this studye
| Parental or mutant (M) strain | MIC (μg/ml) (+ reserpine)a of: |
Mutation |
||||||
|---|---|---|---|---|---|---|---|---|
| NOR | CIP | GAT | MXF | EtBr | GrlA | GyrA | flqB | |
| ATCC 25923 | 2 (1) | 0.5 (0.2) | 0.1 (0.1) | 0.05 (0.05) | 8 (4) | —c | — | — |
| SA-1 | 16 (2) | 16 (0.5) | 0.5 (0.1) | 0.1 (0.05) | 32 (8) | — | — | T→Cd |
| MSA-1 | 2,048 (128) | 256 (64) | 16 (4) | 8 (2) | 32 (8) | Ser80Phe | Ser84Leu | T→Cd |
| Sa2102b | 16 (16) | 2 (2) | 0.2 (0.2) | 0.2 (0.1) | 4 (4) | Ser80Tyr | — | — |
| MSa2102b | 256 (128) | 64 (64) | 8 (4) | 4 (4) | 4 (4) | Ser80Tyr | Ser84Leu | — |
| Sa2667 | 32 (32) | 4 (4) | 0.5 (0.5) | 0.5 (0.5) | 8 (4) | Ser80Phe | — | — |
| MSa2667 | 2,048 (256) | 256 (64) | 8 (4) | 2 (1) | 128 (8) | Ser80Phe | Ser84Ala | — |
| Sa2669b | 64 (32) | 16 (8) | 1 (1) | 0.5 (0.5) | 4 (2) | Ser80Phe | Ser84Ala | — |
| MSa2669b | 128 (128) | 32 (32) | 4 (4) | 2 (2) | 4 (4) | Ser80Phe | Ser84Val | — |
MICs indicated in parentheses have been determined in presence of reserpine at 128 μg/ml.
Methicillin-resistant S. aureus.
—, no mutation.
Eighty-eight bases upstream from the ATG initiation codon of the norA gene.
Abbreviations: NOR, norfloxacin; CIP, ciprofloxacin; GAT, gatifloxacin; MXF, moxifloxacin; EtBr, ethidium bromide.
TABLE 2.
Primers used in this study
| Gene amplification (nucleotide positions) | Primer | Sequence (5′ to 3′) |
|---|---|---|
| grlA (2347 to 2870)a | GrlAF2 | AATGATCAATTTGATGAGGAGG |
| GrlAR2 | CACTAGTAAGTTAGGAAATCT | |
| grlB (1562 to 1877)a | GrlBF2 | GTGAAGATGCTCGTTCAGG |
| GrlBR2 | ACGATTATAATTACTATCTT | |
| gyrA (2152 to 2658)b | GyrAF1 | ATGGCTGAATTACCTCAATCAAG |
| GyrAR1 | GGCTAATAAGTTAGGGAATCGA | |
| gyrB (1379 to 1686)b | GyrBF1 | GTGAAGTAACACGTCGTA |
| GyrBR1 | TTTGTGATATCTTGCTTTCGC | |
| flqB (129 to 546)c | NorAL2 | AACGTCATCACATGCACCA |
| NorAL3 | TATTACTAAACCGATACC |
Operating procedure and antimicrobial assays.
The peripheral compartment (PCp) of the model was inoculated with 30 ml of an overnight suspension for each bacterial strain, 3 h before performing the first sampling, in order to obtain an exponentially growing culture of ∼107 CFU/ml in a 50-ml culture volume. Growth control and killing curves were carried out for ATCC 25923 in duplicate. Bacterial counts were measured only at 0, 24, and 48 h in the presence of gatifloxacin for the other strains.
During the validation tests with the reference strain ATCC 25923, a total of 29 samples of 200 μl each (at 0, and 0.25, 0.5, 0.75, 1, 1.5, 2, 2.5, 3, 4, 6, 8, 12, 18, and 24 h after antibiotic administration for each regimen interval) were drawn from the PCp by manual sampling with sterile Vacutainer tubes (PolyLabo Division of VWR, Strasbourg, France). A 10-μl aliquot of each sample was used for antibiotic dosing by a high-pressure liquid chromatography assay (29). A 100-μl volume from 17 PCp samples (0, 1, 2, 3, 4, 6, 8, 12, and 24 h after antibiotic administration for each regimen interval) was devoted to the bacterial growth quantification. For bacterial counts in the absence of antibiotic, a total of 17 samples were drawn from the PCp at 3, 4, 5, 6, 7, 9, 11, 15, 27, 28, 29, 30, 31, 33, 35, 39, and 51 h after bacterial inoculation. Suspensions were 10-fold-diluted with a sterile 0.9% NaCl solution and plated on MH agar. After incubation for 24 h at 37°C, colonies were enumerated and bacterial counts were calculated and expressed as log10 CFU/milliliter. In the case of treatment simulations, the emergence of resistant mutants was monitored by plating the same diluted samples (at 0, 24, and 48 h after antibiotic administration) on MH agar supplemented with 0.2, 0.5, 1, 2, and 8 μg/ml of gatifloxacin. Bacterial counts were determined by dilution plating as indicated above. In order to avoid carryover effect, the first 10-fold diluted sample was not plated. Consequently, the lower detection and counting limits were 2 and 3 log10 CFU/ml, respectively. For the strains other than ATCC 25923, the number of samples was reduced to four (1, 24, 25, and 48 h) for the antibiotic assay and to five (0, 1, 24, 25, and 48 h) for bacterial counts. All experiments were performed in duplicate.
Data analysis. (i) Pharmacokinetic analysis.
Compartmental analysis of drug experimental concentration data obtained during investigation with the reference strain ATCC 25923 was performed with the software Pharmacokin (G. Kister, J. Bres, and G. Cassanas, Prog. Abstr. 2nd Sci. Meet. Assoc. Pharmacy Faculties Pharmacologists, abstr. 13, p. 14, 1998). The goodness of fit for each concentration-time curve was evaluated by the correlation coefficient between experimental and software-calculated data. The Cmax and the residual concentration at the end of the administration interval (Cres) were taken directly from concentration-time profiles, whereas the t1/2β, the half-life at α phase (t1/2α), the area under the concentration-time curve (AUC) within the different dosing intervals and its extrapolation to infinite, the mean residence time (MRT), total clearance (CLtot), apparent volume of distribution (Vβ), and volume of distribution at steady state (Vss) were calculated. The Cmax and Cres values were controlled during investigations with the other strains.
(ii) Quantification of the bacterial growth and evaluation of the antibacterial effect.
For the reference strain ATCC 25923, the doubling time of the bacterial population was determined by linear regression analysis of the log10 CFU/milliliter versus time during the exponential phase (from 5 to 9 h postinoculation). For the same strain, the MIC-related pharmacokinetic parameters (inhibitory quotient [Cmax/MIC] [8] and AUC divided by the MIC [AUC/MIC] [6, 10, 26]) and the indices of bacterial killing in the presence of antibiotic (bacterial killing and regrowth curve from 0 to 48 h [AUBC0-48], area under the control growth curve from 0 to 48 h [AUGC0-48], area between the control growth curve and the bacterial killing and regrowth curve from the zero point to 48 h [ABBC0-48] [11], and difference between the bacterial counts at the beginning of the treatment and at a defined time [Δlog CFU/milliliter] [6, 25]) were calculated. The Cmax/MIC and the Δlog CFU/milliliter were also determined for the other strains, and the AUC/MIC ratios were calculated using the AUC0-24 reproducibly obtained for the reference strain. The rate of resistant mutants was expressed as the percentage of survivors grown on antibiotic-supplemented agar plates.
RESULTS
Antibiotic assays and pharmacokinetic data.
Fluorescence detection allowed a quantification limit of 0.1 μg/ml for gatifloxacin in MH broth and a good selectivity. The standard curve was linear between 0.1 and 8 μg/ml, with a correlation coefficient of >0.999 (n = 6). The online extraction from a C18 precolumn led to recoveries of 108% ± 7.6%, 105% ± 0.8%, and 106% ± 1.2% for quality control samples of 0.25, 2.5, and 5 μg/ml, respectively. Intraday and interday coefficients of variation within the linearity range varied from 0.57 to 3.61% and from 1.30 to 8.66%, respectively. Intraday and interday accuracies ranged from 2.06 to 5.19% and from −3.68 to 4.56%, respectively. Mean concentration-time curves for the reference strain (data not shown) allowed us to determine pharmacokinetic parameters (Table 3), and the coefficient of correlation between experimental data and the calculated pharmacokinetic profile was always greater than 0.98. Concentrations for the ATCC 25923 strain at the peak (5.36 and 5.80 μg/ml at t1 and t25, respectively) and the trough (0.33 and 0.34 μg/ml for t24 and t48, respectively) were similar to those for the other strains (4.60 to 4.99 and 5.06 to 5.86 μg/ml as well as 0.43 to 0.53 and 0.40 to 0.58 μg/ml, respectively).
TABLE 3.
Mean pharmacokinetic parameters (n = 2) for the peripheral (PCp) compartment after simulation of a once-daily 1-h infusion of 400 mg of gatifloxacin over 48 h and the corresponding human reference data
| Parameter (unit) | Dosing interval |
Human dataa |
||
|---|---|---|---|---|
| 0-24 h | 24-48 h | Female (n = 12) | Male (n = 12) | |
| Cmax (μg/ml) | 5.36 | 5.80 | 6.07 ± 0.77 | 5.47 ± 0.98 |
| Cres (μg/ml) | 0.33 | 0.34 | ||
| t1/2α (h) | 0.40 | 0.49 | ||
| t1/2β (h) | 8.68 | 7.80 | 6.24 ± 0.85 | 7.66 ± 0.83 |
| AUC0-24 (h × μg/ml) | 30.6 | 35 | ||
| AUC0-∞ (h × μg/ml) | 34.7 | 38.9 | 37.4 ± 5.61 | 34.1 ± 8.50 |
| MRT (h) | 11.1 | 10.3 | 8.10 ± 1.28 | 9.41 ± 1.34 |
| CL (liters/h) | 11.5 | 10.3 | 10.9 ± 1.74 | 12.3 ± 2.44 |
| CL (ml/min) | 192 | 171 | 182 ± 28.9 | 205 ± 40.8 |
| Vβ (liters) | 144 | 116 | 97.1 ± 10.7 | 133 ± 18.6 |
| Vss (liters) | 125 | 104 | 87.1 ± 9.33 | 113 ± 17.3 |
| Correlation coefficient | 0.99 | 0.99 | ||
Human data are from LaCreta et al. (24).
Characterization of the bacterial parental strains.
To evaluate the potential efficacy of gatifloxacin in the PK-PD model, in addition to the wild-type reference strain ATCC 25923, four strains (SA-1, Sa2102, Sa2667, and Sa2669) were selected by preliminary experiments. In a first step of screening, clinical strains of S. aureus exhibiting low-level ciprofloxacin resistance by a routine agar diffusion method were found to be infrequent in our collection (e.g., mean data in two hospitals of our region in 2005 for MSSA [64%] and MRSA [36%]: 3 and 6% ciprofloxacin intermediate and 7 and 69% ciprofloxacin resistant). MIC determination for 30 of them showed that 29, displaying ciprofloxacin MICs of 2 to 16 μg/ml, could be considered gatifloxacin susceptible using the same MIC breakpoints (MICs, 0.2 to 1 μg/ml; mode MIC for 22 strains, 0.2 μg/ml). In a second step of screening, sequencing demonstrated that among five strains exhibiting the representative gatifloxacin MICs of 0.2, 0.5, and 1 μg/ml, four had mutations in the QRDR of grlA (Ser80Phe for three, including Sa2667, and Ser80Tyr for Sa2102) or in the QRDR of both grlA and gyrA for the most resistant strain (Ser80Phe and Ser84Ala, respectively, for Sa2669) (Table 1). Since none possessed mutations in the promoter region of norA, SA-1 was selected from the reference strain ATCC 25923 on an EtBr gradient. This strain exhibited, as expected for a NorA-overexpressing mutant, increased MICs of the hydrophilic fluoroquinolones (norfloxacin and ciprofloxacin) and EtBr that were restored by the efflux pump inhibitor reserpine. Moreover, sequencing revealed the presence of a T→C change 88 bp upstream from the ATG initiation codon of norA, while the topoisomerase-encoding genes had the same sequences as the reference strain.
Pharmacodynamic data. (i) Growth control curves.
The initial inoculum of strain ATCC 25923 (3 h after inoculation of the PCp) was 5 × 106 CFU/ml. Exponential growth continued up to 12 h after the initial sampling time (t0). The mean doubling time (n = 2) during the exponential phase was 0.75 h. A plateau at ∼9 log10 CFU/ml was then observed. Thus, bacterial growth was not affected by the experimental conditions. Similar values of the initial inoculum were obtained for the other strains under study (4 × 106 to 9 × 106 CFU/ml).
(ii) Killing curves.
During exposure at the initial dose of the simulation of daily 1-h infusion of 400 mg of gatifloxacin for 48 h of treatment, bacterial count of the strain ATCC 25923 decreased rapidly and was below the limit of detection at 12 h after the beginning of drug administration. No regrowth was noted within the treatment duration. For SA-1, Sa2102, Sa2667, and Sa2669, a 1-log decrease was observed at the end of the 1-h infusion. Bacterial concentrations at 24 h were close to those obtained at 1 h after the beginning of drug administration, and they reached at least 108 CFU/ml at 48 h, indicating the inefficacy of the second dose.
(iii) MIC-related pharmacokinetic parameters and quantitative evaluation of in vitro antibacterial effect.
The Cmax/MIC ratios decreased exponentially from 53.6 to 4.93 at 1 h and from 58.0 to 5.68 at 25 h, leading to the following classification according to the strains' initial susceptibility: ATCC 25923 > Sa2102 > (SA-1 and Sa2667) > Sa2669 (Table 4). The AUC0-24/MIC exhibited the same decreasing profile. The Δlog CFU/milliliter at the end of the first infusion was negative for all the strains. At 24 and 25 h, only the Sa2102 strain gave slightly positive values for this parameter, despite its low initial MIC (Table 1). At 48 h, except for ATCC 25923, all the strains under study exhibited positive values for Δlog CFU/milliliter. With ATCC 25923, the calculated AUC/MIC ratio for a 24-h interval was 306 and ranged from 30.6 to 153 with the other strains. The rapid eradication of this strain led to a low AUBC0-48 and an ABBC0-48 (419 h × log CFU/milliliter) close to the AUGC0-48 (449 h × log CFU/milliliter).
TABLE 4.
Pharmacodynamic values determined for the five tested strains of S. aureus (n = 2)
| Parameter and time point | Pharmacodynamic value for strain: |
||||
|---|---|---|---|---|---|
| ATCC 25923 (MIC = 0.1 μg/ml) | SA-1 (MIC = 0.5 μg/ml) | Sa2102 (MIC = 0.2 μg/ml) | Sa2667 (MIC = 0.5 μg/ml) | Sa2669 (MIC = 1.0 μg/ml) | |
| Cmax/MIC | |||||
| 1 h | 53.6 | 9.76 | 24.9 | 9.20 | 4.93 |
| 25 h | 58.0 | 10.1 | 29.3 | 10.2 | 5.68 |
| AUC0-24/MIC | 306 | 61.2 | 153 | 61.2 | 30.6 |
| Δlog CFU/ml | |||||
| t1 | −2.18 | −1.22 | −0.59 | −0.49 | −0.33 |
| t24 | −6.80 | −1.80 | 0.04 | −0.92 | −0.43 |
| t25 | −6.80 | −1.68 | 0.05 | −1.43 | −0.58 |
| t48 | −6.80 | 1.53 | 3.02 | 2.57 | 2.25 |
| ABBC0-48 (h × log CFU/ml) | 419 | ||||
(iv) Emergence of resistant mutants.
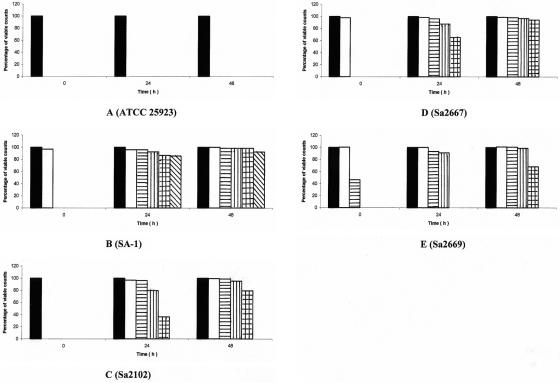
With the reference strain ATCC 25923, no survivors able to grow in the presence of gatifloxacin concentrations equal or superior to the MIC for the parental strains emerged (Fig. 1A). However, with the other strains, survivors developed in the presence of 4-fold (Sa2667), 10-fold (Sa2102), and 16-fold (SA-1) the gatifloxacin MIC for the parental strain at the 24th hour. Slightly higher rates of survival were observed in the presence of the same concentrations at the 48th hour, when survivors appeared on twice the gatifloxacin MIC for the parental strain Sa2669 (Fig. 1B to E). For each strain, three independent clones were selected at the highest antibiotic concentrations at 48 h. They gave the same antibiotic resistance profile, identical to the parental strain except for high-level ciprofloxacin resistance by the agar diffusion method (data not shown), and gatifloxacin MICs that were superior to 2 μg/ml (Table 1).
FIG. 1.
Emergence of gatifloxacin-resistant mutants during simulation of once-daily 1-h infusion of 400 mg over 48 h in the in vitro PK-PD model on S. aureus ATCC 25923 (A), its mutant SA-1 (B), and the clinical strains Sa2102 (C), Sa2667 (D), and Sa2669 (E) (mean, n = 2). Bars: ▪, control; □, resistant to 0.2 μg/ml; ▤, resistant to 0.5 μg/ml; ▥, resistant to 1.0 μg/ml; ▩, resistant to 2.0 μg/ml; ▧, resistant to 8.0 μg/ml.
Molecular analysis was then conducted for a single clone of each series of survivors (Table 1). The MSA-1 mutant, which was 32 times more resistant to gatifloxacin than its parental strain, exhibited two supplementary substitutions in the grlA (Ser80Phe) and the gyrA (Ser84Leu) QRDRs. The MSa2102 mutant, 40-fold more resistant to gatifloxacin than Sa2102, possessed an additional Ser84Leu mutation in the gyrA QRDR. The MSa2667 mutant, leading to a 16-fold increase in MIC compared to that of Sa2667, also harbored an additional mutation in gyrA (Ser84Ala). Furthermore, a higher increase in norfloxacin and ciprofloxacin MICs (64-fold and 16- to 32-fold, respectively) and an increase in EtBr MIC were observed in MSa2667. These increases were significantly reduced by the addition of reserpine, suggesting the overproduction of an efflux system, although there were no mutations in the 331-bp region located upstream from the norA gene. Finally, the MSa2669 mutant, fourfold more resistant than Sa2669, presented a change in the Ser84 mutation in GyrA, from Ala to Val.
DISCUSSION
New fluoroquinolones, such as gatifloxacin, have been primarily developed for treating community-acquired pulmonary infections due to multiresistant pneumococci. However, their enhanced in vitro activity against staphylococci (17, 18) argue for their use rather than the older fluoroquinolones, such as ciprofloxacin, in the treatment of staphylococcal infections. However, this assumption warrants further investigations.
In this study, we examined the activity of gatifloxacin in a PK-PD model mimicking the plasma fluctuation concentrations. These models are known to afford more clinically reliable information than static measures of MICs (1, 3, 5, 15). Since gatifloxacin binding by serum proteins is approximately 20% independent of concentrations (12), and since protein binding is a modest contributor to the therapeutic effectiveness of fluoroquinolones (4), non-protein-supplemented broth was used to simulate human total (free plus bound) concentrations of the drug. After validation of the bacterial growth and of the antibiotic assay, simulation of the recommended treatment of 1-h-daily infusion of 400 mg for 48 h provided pharmacokinetic parameters highly similar to those from human data (24).
The pharmacodynamic effect of gatifloxacin was first evaluated against the reference strain of S. aureus ATCC 25923. Due to a high intrinsic activity (MIC, 0.1 μg/ml), the concentration/MIC ratios of gatifloxacin reached 53 to 58 at the Cmax and were still 3 to 4 at the Cres at 24 h. Since gatifloxacin is, like other quinolones, a zwitterion (pKa1 6.0 and pKa2 9.2) with an isoelectric point at pH 7.7, the degree of ionization is low in plasma (pH 7.4) (27). According to the principle of nonionic diffusion and to the low protein binding of fluoroquinolones, the highly vascularized tissue concentration of such a substance can be expected to approximate plasma levels such as those simulated in our model. At therapeutic plasma concentrations, gatifloxacin led to the complete eradication of the sensitive strain ATCC 25923 within 12 h after the beginning of the first infusion and to a low value of AUBC0-48 (30.2 h × log CFU/milliliter). This high bactericidal effect is in good agreement with the high values of the inhibitor quotient of Cmax/MIC and of AUC0-24/MIC. Gatifloxacin is known to select less frequently bacterial resistant mutants than older fluoroquinolones (19). This property seems to be due to the inhibition of both DNA gyrase and topoisomerase IV at nearly the same level (14, 32), while most earlier fluoroquinolones recognize topoisomerase IV as a primary target in S. aureus (28). This dual-targeting capacity of gatifloxacin would be related to the presence of a C8-methoxy group, which also explains its better activity (7, 19). In addition, owing to its high hydrophobicity (logP, +0.69), gatifloxacin is a poor substrate for the main multidrug resistance efflux pump of S. aureus NorA and is not expected to easily select efflux-resistant mutants (19).
The pharmacodynamic effect of gatifloxacin was then evaluated on four gatifloxacin-susceptible strains of S. aureus exhibiting various levels and mechanisms of ciprofloxacin resistance. The preliminary screening experiments performed to select these strains showed that while ciprofloxacin susceptibility is much more common among MSSA than MRSA, low-level resistance is equally infrequent in both populations. Sequencing revealed that the selected clinical strains harbored the usual target mutations (31, 35), known to confer MICs exceeding the susceptibility breakpoint of ciprofloxacin but not of gatifloxacin (16, 19). Efflux mutants were not recovered from the tested clinical strains. Indeed, overexpression of efflux pumps is essentially found in S. aureus strains highly resistant to ciprofloxacin that also carry target mutations (30). Thus, SA-1, an overexpressing NorA mutant of S. aureus ATCC 25923, was selected in vitro. Sequencing of the norA promoter region revealed a base transition at position −88, consistent with an overproduction of NorA (13, 23), reported here for the first time.
For these four selected strains, the Cmax/MIC (4.93 to 29.3) and the AUC0-24 h/MIC (30.6 to 153) were substantially lower than the values obtained with S. aureus ATCC 25923. Accordingly, regrowth occurred, and bacterial concentrations close to the initial inoculum were observed 24 h after the beginning of the first infusion. The Δlog CFU/ml ranged from −1.80 to 0.04. The second dose had no appreciable effect for all strains, and bacterial counts close to the plateau value were reached at 48 h. Bacterial regrowth was associated with acquired antibiotic resistance, since the surviving cells were capable of growing on media containing multiples of gatifloxacin MICs for the initial strains. Antibiotic susceptibility testing and MIC determination, after subculture on antibiotic-free media, confirmed that these survivors were resistant mutants with MICs ranging between 4 and 16 μg/ml rather than transiently induced cells or persisters. Thus, even simulating total gatifloxacin concentrations for the experiments with the isolates with low-level ciprofloxacin resistance did not hinder the emergence of resistance.
Sequencing demonstrated that the mutants acquired mutations either supplementary (MSa2102 and MSa2667) or different (Ala84Val for MSa2669) in gyrA or in both gyrA and grlA QRDRs (MSA-1). The increases in MICs were related not only to the number but also to the type of amino acid changes. For example, the level of resistance associated with the substitution of Ser-84 in GyrA by nonpolar amino acids is known to be correlated with the length of their aliphatic chains and their hydrophobicities, i.e., Leu conferring higher MICs than Val, itself conferring higher MICs than Ala (33). Furthermore, the presence of efflux systems also interferes. For MSa2667, the overproduction of an efflux pump(s) was not associated with a mutation in the norA promoter region. However, other regulatory mutations leading to the overexpression of NorA, such as in the mgrA or the arlRS loci, or mutations yielding an overproduction of other MDR efflux pump(s), such as the recently described MepA or NorB system, might be implicated (13, 21, 23, 34). On the other hand, the capability of SA-1 to generate a mutant with high-level resistance and cumulative target-based mutations can be ascribed to the fact that efflux-related resistance favors this emergence by diminishing intracellular drug concentrations (23).
In conclusion, our data highlight the beneficial pharmacokinetic and pharmacodynamic characteristics of gatifloxacin that allowed the prevention of the emergence of resistant mutants from a susceptible control strain of S. aureus. Thus, they confirm that gatifloxacin might be preferred to older fluoroquinolones in the treatment of staphylococcal infections due to ciprofloxacin-susceptible strains, which are widespread among MSSA. However, our data also support the idea that fluctuation of plasma concentrations of gatifloxacin alone possess the potential for selecting resistant mutants from strains already presenting various levels of resistance to older fluoroquinolones, similarly encountered among MSSA and MRSA, and whatever the mechanisms are. These findings indicate that the novel fluoroquinolones should be used to treat staphylococcal infections after antibiotic susceptibility testing of both older and novel drugs of this class for MSSA and MRSA in the community as in the health care setting.
Acknowledgments
We thank Catherine André and Laure Coulange for technical assistance and Isabelle Maachi-Guillot (Haut Levêque Hospital Pharmacy Service) and Sylvie Darmagnac (Fresenius Medical Care) for providing disposable devices.
This work was supported in part by a grant of the Ministère de l'Education Nationale et de la Recherche and of Grünenthal Laboratories.
REFERENCES
- 1.Allen, G. P., G. W. Kaatz, and M. J. Rybak. 2003. Activities of mutant prevention concentration-targeted moxifloxacin and levofloxacin against Streptococcus pneumoniae in an in vitro pharmacodynamic model. Antimicrob. Agents Chemother. 47:2606-2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ba, B. B., A. Bernard, A. Iliadis, C. Quentin, D. Ducint, R. Etienne, M. Fourtillan, I. Maachi-Guillot, and M. C. Saux. 2001. New approach for accurate simulation of human pharmacokinetics in an in vitro pharmacodynamic model: application to ciprofloxacin. J. Antimicrob. Chemother. 47:223-227. [DOI] [PubMed] [Google Scholar]
- 3.Bergan, T., I. B. Carlsen, and J. E. Fuglesang. 1980. An in vitro model for monitoring bacterial responses to antibiotic agents under simulated in vivo conditions. Infection 8(Suppl. 1):S96-S102. [DOI] [PubMed]
- 4.Bergogne-Berezin, E. 2002. Clinical role of protein binding of quinolones. Clin. Pharmacokinet. 41:741-750. [DOI] [PubMed] [Google Scholar]
- 5.Booker, B. M., P. F. Smith, A. Forrest, J. Bullock, P. Kelchlin, S. M. Bhavnani, R. N. Jones, and P. G. Ambrose. 2005. Application of an in vitro infection model and simulation for reevaluation of fluoroquinolone breakpoints for Salmonella enterica serotype typhi. Antimicrob. Agents Chemother. 49:1775-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowker, K. E., M. Wootton, C. A. Rogers, R. Lewis, H. A. Holt, and A. P. MacGowan. 1999. Comparison of in-vitro pharmacodynamics of once and twice daily ciprofloxacin. J. Antimicrob. Chemother. 44:661-667. [DOI] [PubMed] [Google Scholar]
- 7.Dong, Y., X. Zhao, J. Domagala, and K. Drlica. 1999. Effect of fluoroquinolone concentration on selection of resistant mutants of Mycobacterium bovis BCG and Staphylococcus aureus. Antimicrob. Agents Chemother. 43:1756-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellner, P. D., and H. C. Neu. 1981. The inhibitory quotient. A method for interpreting minimum inhibitory concentration data. JAMA 246:1575-1578. [DOI] [PubMed] [Google Scholar]
- 9.Ferrero, L., B. Cameron, and J. Crouzet. 1995. Analysis of gyrA and grlA mutations in stepwise-selected ciprofloxacin-resistant mutants of Staphylococcus aureus. Antimicrob. Agents Chemother. 39:1554-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Firsov, A. A., A. A. Shevchenko, S. N. Vostrov, and S. H. Zinner. 1998. Inter- and intraquinolone predictors of antimicrobial effect in an in vitro dynamic model: new insight into a widely used concept. Antimicrob. Agents Chemother. 42:659-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Firsov, A. A., S. N. Vostrov, A. A. Shevchenko, and G. Cornaglia. 1997. Parameters of bacterial killing and regrowth kinetics and antimicrobial effect examined in terms of area under the concentration-time curve relationships: action of ciprofloxacin against Escherichia coli in an in vitro dynamic model. Antimicrob. Agents Chemother. 41:1281-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fish, D. N., and D. S. North. 2001. Gatifloxacin, an advanced 8-methoxy fluoroquinolone. Pharmacotherapy 21:35-59. [DOI] [PubMed] [Google Scholar]
- 13.Fournier, B., R. Aras, and D. C. Hooper. 2000. Expression of the multidrug resistance transporter NorA from Staphylococcus aureus is modified by a two-component regulatory system. J. Bacteriol. 182:664-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukuda, H., S. Hori, and K. Hiramatsu. 1998. Antibacterial activity of gatifloxacin (AM-1155, CG5501, BMS-206584), a newly developed fluoroquinolone, against sequentially acquired quinolone-resistant mutants and the norA transformant of Staphylococcus aureus. Antimicrob. Agents Chemother. 42:1917-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gumbo, T., A. Louie, M. R. Deziel, and G. L. Drusano. 2005. Pharmacodynamic evidence that ciprofloxacin failure against tuberculosis is not due to poor microbial kill but to rapid emergence of resistance. Antimicrob. Agents Chemother. 49:3178-3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hooper, D. C. 2002. Fluoroquinolone resistance among gram-positive cocci. Lancet Infect. Dis. 2:530-538. [DOI] [PubMed] [Google Scholar]
- 17.Hosaka, M., T. Yasue, H. Fukuda, H. Tomizawa, H. Aoyama, and K. Hirai. 1992. In vitro and in vivo antibacterial activities of AM-1155, a new 6-fluoro-8-methoxy quinolone. Antimicrob. Agents Chemother. 36:2108-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ince, D., R. Aras, and D. C. Hooper. 1999. Mechanisms and frequency of resistance to gatifloxacin in comparison with ciprofloxacin in Staphylococcus aureus. Drugs 58(Suppl. 2):134-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ince, D., and D. C. Hooper. 2001. Mechanisms and frequency of resistance to gatifloxacin in comparison to AM-1121 and ciprofloxacin in Staphylococcus aureus. Antimicrob. Agents Chemother. 45:2755-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito, H., H. Yoshida, M. Bogaki-Shonai, T. Niga, H. Hattori, and S. Nakamura. 1994. Quinolone resistance mutations in the DNA gyrase gyrA and gyrB genes of Staphylococcus aureus. Antimicrob. Agents Chemother. 38:2014-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaatz, G. W., F. McAleese, and S. M. Seo. 2005. Multidrug resistance in Staphylococcus aureus due to overexpression of a novel multidrug and toxin extrusion (MATE) transport protein. Antimicrob. Agents Chemother. 49:1857-1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaatz, G. W., S. M. Seo, and C. A. Ruble. 1993. Efflux-mediated fluoroquinolone resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 37:1086-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaatz, G. W., R. V. Thyagarajan, and S. M. Seo. 2005. Effect of promoter region mutations and mgrA overexpression on transcription of norA, which encodes a Staphylococcus aureus multidrug efflux transporter. Antimicrob. Agents Chemother. 49:161-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LaCreta, F. P., S. Kaul, G. D. Kollia, G. Duncan, D. M. Randall, and D. M. Grasela. 2000. Interchangeability of 400-mg intravenous and oral gatifloxacin in healthy adults. Pharmacotherapy 20:59S-66S. [DOI] [PubMed] [Google Scholar]
- 25.MacGowan, A. P., K. E. Bowker, M. Wootton, and H. A. Holt. 1999. Exploration of the in-vitro pharmacodynamic activity of moxifloxacin for Staphylococcus aureus and Streptococci of lancefield groups A and G. J. Antimicrob. Chemother. 44:761-766. [DOI] [PubMed] [Google Scholar]
- 26.Madaras-Kelly, K. J., B. E. Ostergaard, L. B. Hovde, and J. C. Rotschafer. 1996. Twenty-four-hour area under the concentration-time curve/MIC ratio as a generic predictor of fluoroquinolone antimicrobial effect by using three strains of Pseudomonas aeruginosa and an in vitro pharmacodynamic model. Antimicrob. Agents Chemother. 40:627-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naber, C. K., M. Steghafner, M. Kinzig-Schippers, C. Sauber, F. Sorgel, H. J. Stahlberg, and K. G. Naber. 2001. Concentrations of gatifloxacin in plasma and urine and penetration into prostatic and seminal fluid, ejaculate, and sperm cells after single oral administrations of 400 milligrams to volunteers. Antimicrob. Agents Chemother. 45:293-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ng, E. Y., M. Trucksis, and D. C. Hooper. 1996. Quinolone resistance mutations in topoisomerase IV: relationship to the flqA locus and genetic evidence that topoisomerase IV is the primary target and DNA gyrase is the secondary target of fluoroquinolones in Staphylococcus aureus. Antimicrob. Agents Chemother. 40:1881-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen, H. A., J. Grellet, B. B. Ba, C. Quentin, and M. C. Saux. 2004. Simultaneous determination of levofloxacin, gatifloxacin and moxifloxacin in serum by liquid chromatography with column switching. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 810:77-83. [DOI] [PubMed] [Google Scholar]
- 30.Schmitz, F. J., A. C. Fluit, S. Brisse, J. Verhoef, K. Kohrer, and D. Milatovic. 1999. Molecular epidemiology of quinolone resistance and comparative in vitro activities of new quinolones against European Staphylococcus aureus isolates. FEMS Immunol. Med. Microbiol. 26:281-287. [DOI] [PubMed] [Google Scholar]
- 31.Schmitz, F. J., M. E. Jones, B. Hofmann, B. Hansen, S. Scheuring, M. Luckefahr, A. Fluit, J. Verhoef, U. Hadding, H. P. Heinz, and K. Kohrer. 1998. Characterization of grlA, grlB, gyrA, and gyrB mutations in 116 unrelated isolates of Staphylococcus aureus and effects of mutations on ciprofloxacin MIC. Antimicrob. Agents Chemother. 42:1249-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takei, M., H. Fukuda, R. Kishii, and M. Hosaka. 2001. Target preference of 15 quinolones against Staphylococcus aureus, based on antibacterial activities and target inhibition. Antimicrob. Agents Chemother. 45:3544-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takenouchi, T., C. Ishii, M. Sugawara, Y. Tokue, and S. Ohya. 1995. Incidence of various gyrA mutants in 451 Staphylococcus aureus strains isolated in Japan and their susceptibilities to 10 fluoroquinolones. Antimicrob. Agents Chemother. 39:1414-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Truong-Bolduc, Q. C., P. M. Dunman, J. Strahilevitz, S. J. Projan, and D. C. Hooper. 2005. MgrA is a multiple regulator of two new efflux pumps in Staphylococcus aureus. J. Bacteriol. 187:2395-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, T., M. Tanaka, and K. Sato. 1998. Detection of grlA and gyrA mutations in 344 Staphylococcus aureus strains. Antimicrob. Agents Chemother. 42:236-240. [DOI] [PMC free article] [PubMed] [Google Scholar]