Abstract
Resistance of clinical isolates of Candida albicans to the echinocandin drug caspofungin is slowly emerging and is linked to mutations in short conserved regions in the FKS1 gene. The most prominent changes occurred at the serine 645 position in Fks1p with substitutions of proline, tyrosine, and phenylalanine. An allele-specific real-time PCR molecular-beacon assay was developed for rapid identification of drug resistance by targeting FKS1 mutations. Mutations altering serine 645 were reliably identified in both heterozygous and homozygous states. The molecular-beacon assay was used to evaluate two large collections of spontaneous mutants from separate strains of C. albicans with resistance (MICs, >16 μg/ml) to caspofungin with the goal of understanding the relationship between FKS1 mutations and echinocandin resistance. Of 85 resistant isolates recovered, all were identified with mutations in FKS1; 93% showed changes at Ser645, with 62% displaying a characteristic S645P substitution expressed as either a homozygous or a heterozygous mutation in FKS1. Two other prominent amino acid substitutions, S645Y and S645F, were found at frequencies of 22% and 8%, respectively. Three new mutations were also identified: T1922C, G1932T, and C1934G, encoding F641S, L644F, and S645C substitutions, respectively. One strain had the double amino acid substitution L644F and S645C. Allele-specific probes were combined in a multiplex assay for reliable screening of known FKS1 mutations. These data support the importance of FKS1p substitutions in echinocandin resistance and demonstrate the feasibility of applying molecular screening for routine resistance assessment.
The echinocandin drugs, caspofungin, micafungin, and anidulafungin, are the first of a new class of antifungal compounds that target the fungal cell wall by blocking β-1,3-glucan synthase (4, 6, 9). These drugs have broad-spectrum activity against Candida and Aspergillus spp. without cross-resistance to existing agents and therefore are effective against azole-resistant yeasts and molds (1, 7, 15, 18, 19). Importantly, due to their critical effect on the cell wall, echinocandins kill yeasts (5). Use of caspofungin (CANCIDAS), the first approved echinocandin, in the clinic is expanding rapidly, and it is now widely used along with triazole drugs, such as voriconazole, for primary therapy against yeast and molds. The entry of the closely related drugs micafungin and anidulafungin will further extend the clinical scope of this highly efficacious class of drugs. As patient exposure to caspofungin grows, and as micafungin and anidulafungin augment the market, it is anticipated that the number of clinical isolates with highly elevated MICs will increase. Because the echinocandins are the first new major antifungal drug class to enter the market in decades, it is vital to assess the nature of developing resistance mechanisms to this class of drugs.
Recently, we reported that specific mutations in two highly conserved regions of the Fks1p subunit of glucan synthase, a putative large polytopic membrane protein, can confer resistance to caspofungin in laboratory and clinical isolates of Candida, as defined by elevated MICs, a decrease of several orders of magnitude in the sensitivity of mutant enzymes to the drug, and a comparable shift in sensitivity to the drug in animal models (16). Mutations affecting the target site are the most likely mechanism of resistance, since, unlike azole antifungal drugs, the echinocandins are poor substrates for drug efflux transporters (1, 24). High-level reduced susceptibility to caspofungin among clinical isolates is associated with amino acid substitutions in two regions of Fks1p (16). Most missense mutations affect the same FKS1 locus, leading to substitutions of serine 645 for proline, phenylalanine, and tyrosine. The mutations were dominant, displaying phenotypic resistance when present in both homozygous and heterozygous forms. To further evaluate the contribution of mutations in FKS1 as a resistance mechanism, we have investigated extensive collections of spontaneous mutants resistant to caspofungin. To help in these studies, we have applied nucleic acid-based molecular-beacon technology (26) to develop an allele-specific real-time PCR assay capable of rapid identification of caspofungin resistance mutations in FKS1. The assay allowed fast and reliable identification of both homozygous and heterozygous mutations in FKS1 affecting serine 645. In addition, new mutations in FKS1 were discovered. Finally, the allele-specific probes were combined in a multiplex real-time PCR assay which holds promise for rapid detection of echinocandin resistance.
MATERIALS AND METHODS
Strains, culture conditions, and susceptibility testing.
Candida albicans strains SC5314 (8) and M70 were from the Merck culture collection (MRL, Rahway, NJ). Strains were grown on yeast extract-peptone-dextrose (YPD) medium (1% yeast extract, 2% Bacto Peptone, 2% dextrose). Caspofungin (Merck, Rahway, NJ) was added directly to YPD at 4 μg/ml. Agar plates were incubated at 30°C, and liquid cultures were grown in 14-ml culture tubes containing 3 ml of YPD on a rotary shaker (100 rpm) at 30°C. Susceptibility to caspofungin was estimated by a liquid broth microdilution assay in RPMI 1640 medium (Sigma-Aldrich, St. Louis, MO), as outlined in CLSI (formerly NCCLS) document M27-A2 (14) and described previously (2).
Isolation of caspofungin-resistant C. albicans mutants.
Spontaneous caspofungin-resistant mutants of C. albicans strains SC5314 and M70 were isolated by plating 100 μl (∼108 cells) of an 18-h liquid YPD culture onto YPD plates containing 4 μg/ml caspofungin. Serial dilutions of the overnight cultures were plated onto the YPD plates without antibiotic selection to determine starting colony counts. Selection plates were incubated for 10 to 14 days at 30°C. From each selection plate, at least four individual colonies were reinoculated on fresh caspofungin-containing plates to confirm the resistant phenotype. One truly resistant derivative (≥4 μg/ml) from each culture/plate was used for further studies.
Design of molecular beacons and primers.
Two DNA sequences reported for FKS1 were used for design of FKS1 molecular beacons and primers: GenBank accession number AF027295 and Candida albicans genome database (http://genolist.pasteur.fr/CandidaDB/) accession number CA2043. Nucleotide bases were numbered from the first nucleotide of the FKS1 open reading frame. Molecular beacons and oligonucleotide primers (Table 1) were designed using Beacon Designer software (version 3.0; PREMIER Biosoft, Palo Alto, CA). The default software parameters were applied for construction of all molecular beacons and primers. Molecular beacons were labeled with fluorophores 5-carboxyfluorescein (FAM) and 6-carboxy-2′,4,4′,5′,7,7′-hexachlorofluorescein (HEX) at the 5′ end and with the nonfluorescent quencher 4-{[−4-(dimethylamino)-phenyl]-azo}-benzoic acid (dabcyl) at the 3′ end. Both molecular beacons and primers were purchased from Biosearch Technologies (Novato, CA). Hybridization properties for the FKS1 allele-specific molecular beacons were tested for the full temperature range, 25°C to 95°C, with single-stranded target oligonucleotides (Table 2). Molecular-beacon-target hybridization was performed with the Stratagene (La Jolla, CA) MX4000 multiplex quantitative PCR system. The “molecular-beacon melting curve” experiment type in the MX4000 software was chosen for data monitoring and analysis. Each 50-μl hybridization reaction mixture contained 1× Stratagene Core PCR buffer, 4 mM MgCl2, 100 pmol of the individual target oligonucleotide, and 5 pmol of the molecular beacon. The thermal conditions of the experiment comprised heating at 95°C for 3 min and cooling to 80°C with subsequent cooling down to 25°C using 112 30-s steps with a temperature gradient of −0.5°C. Fluorescence output for each individual reaction was measured at the end of the cooling step. The final data of the “molecular-beacon melting curve” experiment were converted to a “SYBR green (with dissociation curve)” type of experiment. The melting temperature (Tm) for each molecular-beacon-target pair was determined by MX4000 software as a temperature point corresponding to the maximal value of the first derivative of the fluorescence output [−R′(T)]. Out of two Tm values obtained for each pair, the higher Tm value corresponding to more stable beacon-target hybrid with one or no mismatches was taken into account. Each thermal profiling experiment was performed in triplicate.
TABLE 1.
Primer and probe sequences used in this work
| Oligonucleotide | Sequencea,b | 5′ Modification | 3′ Modification | Purpose |
|---|---|---|---|---|
| FKS1-F1719 | CATTGCTGTGGCCACTTTAG | None | None | Sequencing primer |
| FKS1-R2212 | GATTTCCATTTCCGTGGTAGC | None | None | Sequencing primer |
| HS1SN2 | GCCAAATTGGTTGAATCTTA | None | None | Real-time PCR primer |
| HS1AN2 | GTCATGGTCGACAAGTTTCT | None | None | Real-time PCR primer |
| T-WT | AAAAATCTCTTAAAGACAAAGTCAAGAAAAAA | None | None | Wild-type target |
| T-T1933C | AAAAATCTCTTAAAGGCAAAGTCAAGAAAAA | None | None | T1933C mutation target |
| T-C1934A | AAAAATCTCTTAAATACAAAGTCAAGAAGAAAA | None | None | C1934A mutation target |
| T-C1934T | AAAAATCTCTTAAAAACAAAGTCAAGAAGAAAA | None | None | C1934T mutation target |
| T-T1929A | AAAAATCTCTTAAAGACAATGTCAAGAAAAAA | None | None | T1929A SNP target |
| T-T1929A-T1933C | AAAAATCTCTTAAAGGCAATGTCAAGAAAAA | None | None | T1929A and T1933C mutation target |
| T-T1929A-C1934A | AAAAATCTCTTAAATACAATGTCAAGAAGAAAA | None | None | T1929A and C1934A mutation target |
| T-T1929A-C1934T | AAAAATCTCTTAAAAACAATGTCAAGAAGAAAA | None | None | T1929A and C1934T mutation target |
| FKS1(WT) | CGCGAGTTCTTGACWTTGTCTTTAAGAGATCTCGCG | FAM | Dabcyl | Wild-type probe |
| FKS1(T1933C) | CGCGAGTCTTGACWTTGCCTTTAAGAGATCTCGCG | HEX | Dabcyl | T1933C mutation probe |
| FKS1(C1934A) | CGCGAGCTTCTTGACWTTGTATTTAAGAGATCTCGCG | HEX | Dabcyl | C1934A mutation probe |
| FKS1(C1934T) | CGCGAGCTTCTTGACWTTGTTTTTAAGAGATCTCGCG | HEX | Dabcyl | C1934T mutation probe |
Probe domains of molecular beacons and target domains of targets are underlined. Bases in sequences of molecular beacons and targets corresponding to mutations in the C .albicans FKS1gene are boldfaced. All sequences are listed in the 5′ direction.
Each of the degenerate probes represents an equimolar mixture of two identical molecular beacons with either A or T at position W.
TABLE 2.
Melting temperatures of FKS1 molecular beaconsa
| Molecular beacon |
Tm with the following oligonucleotide:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| T-WT | T-T1933C | T-C1934A | T-C1934T | T-T1929A | T-T1929A-T1933C | T-T1929A-C1934A | T-T1929A-C1934T | |
| FKS1(WT) | 63.2 ± 0.5 | 60.4 ± 0.3 | 50.7 ± 0.5 | 51.4 ± 0.3 | 63.0 ± 0.3 | 60.4 ± 0.3 | 51.7 ± 0.5 | 51.9 ± 0.3 |
| FKS1(T1933C) | 52.4 ± 0.3 | 64.0 ± 0.3 | 40.5 ± 0.6 | 40.2 ± 0.0 | 51.5 ± 0.6 | 63.4 ± 0.3 | 39.0 ± 0.3 | 39.5 ± 0.6 |
| FKS1(C1934A) | 60.0 ± 0.3 | 57.2 ± 0.0 | 63.0 ± 0.3 | 57.2 ± 0.0 | 58.9 ± 0.3 | 56.2 ± 0.0 | 62.7 ± 0.5 | 56.0 ± 0.8 |
| FKS1(C1934T) | 59.4 ± 0.3 | 57.4 ± 0.3 | 57.5 ± 0.3 | 64.0 ± 0.3 | 58.9 ± 0.3 | 56.0 ± 0.3 | 57.2 ± 0.0 | 63.7 ± 0.0 |
Melting temperatures of complementary molecular-beacon-target hybrids are boldfaced.
DNA extraction, PCR amplification, and DNA sequencing.
C. albicans chromosomal DNA was extracted from cells grown overnight in liquid YPD medium with the Q-Biogene (Irvine, CA) FastDNA kit. PCR experiments were performed on an iCycler thermocycler (Bio-Rad Laboratories, Hercules, CA). The FKS1 HS1 region was amplified using primers FKS1-F1719 and FKS1-R2212 (Table 1). Each 100-μl PCR mixture contained 0.25 μM of each primer, 2.5 U of iTaq DNA polymerase (Bio-Rad Laboratories, Hercules, CA), 0.5 mM deoxynucleoside triphosphates, 50 mM KCl, 4 mM MgCl2, 20 mM Tris-HCl, pH 8.4, and about 50 ng of C. albicans chromosomal DNA. The cycling conditions were as follows:1 cycle of 3 min at 95°C; 35 cycles of 30 s at 95°C, 30 s at 55°C, and 1 min at 72°C; and 1 cycle of 3 min at 72°C. PCR products were purified using the Montage PCR purification kit (Millipore, Bedford, MA). PCR products for sequencing were obtained and purified with a CEQ dye terminator cycle sequencing Quick Start kit (Beckman Coulter, Fullerton, CA) according to the manufacturer's recommendations by using an iCycler thermal cycler. Primers FKS1-F1719 or FKS1-R2212 were used for the sequencing reaction. The cycling conditions for sequencing PCR were as follows:1 cycle of 3 min at 95°C and 30 cycles of 20 s at 96°C, 20 s at 50°C, and 1 min at 60°C. All DNA sequencing was performed on a CEQ 8000 genetic analysis system (Beckman Coulter, Fullerton, CA). CEQ 8000 genetic analysis system software was used for hardware control as well as for post-run analysis of sequencing results.
Real-time PCR.
Real-time PCR experiments were performed on a Stratagene MX4000 multiplex quantitative PCR system using the “quantitative PCR (multiple standards)” setting. Reagents from the Brilliant QPCR Core Reagent kit (Stratagene, La Jolla, CA) were used for all reactions. Each 50-μl PCR mixture contained 1× Stratagene Core PCR buffer, 0.2 μM of molecular beacon, 0.25 μM each of primers HS1AN2 and HS1SN2 (Table 1), 2.5 U of SureStart Taq DNA polymerase, 0.4 mM deoxynucleoside triphosphates, 4 mM MgCl2, and about 50 ng of C. albicans chromosomal DNA. In multiplex PCR experiments, the concentration of each of the four molecular beacons (Table 1) was 0.2 μM. Real-time PCR thermal cycler parameters were as follows: 1 cycle of 10 min at 95°C and 45 cycles of 30 s at 95°C, 30 s at 61°C, and 30 s at 72°C. The filter gain set of the MX4000 system was changed to FAM-960 HEX-720 with an aim of equalization of the fluorescence signal magnitudes from different molecular beacons. The fluorescence was measured three times during the annealing step. The conditions for multiplex real-time PCR experiments were identical to those for real-time PCR with individual molecular beacons, with the single exception of an annealing temperature of 60°C. A FAM fluorescence channel was assigned for individual reactions with molecular beacon FKS1(WT), whereas a HEX fluorescence channel was used in PCRs with molecular beacons FKS1(T1933C), FKS1(C1934A), and FKS1(C1934T). Fluorescence in multiplex real-time PCRs were monitored by both FAM and HEX optical channels of MX4000 hardware.
Data processing.
Fluorescence signals coming from the Stratagene MX4000 System during PCR amplification were monitored using MX4000 software in real time. At the end of each run, the amplification plot data were converted to a graphic format and stored as image files or exported into Microsoft Office Excel and stored as spreadsheet files. In the case of multiplex PCRs, the final results of PCR amplifications were converted from a “quantitative PCR (multiple standards)” type of experiment to the “quantitative plate read” type of experiment. Total changes in fluorescence for individual fluorophores (Rpost − Rpre) were taken as values for analysis. Results were converted to a graphic or numerical format and stored as image or spreadsheet files.
RESULTS
Design of allele-specific probes for FKS1 mutations.
DNA sequence analysis of FKS1 from more than 50 C. albicans clinical and laboratory isolates with reduced susceptibility to caspofungin revealed three prominent mutations, T1933C, C1934A, and C1934T, resulting in amino acid changes S645P, S645Y, and S645F, respectively (16). Beside those nucleotide substitutions, alignment of sequencing data disclosed another point of variability in this region. In about 25% of all C. albicans strains analyzed (n > 100), including genome reference strain SC5314 (8), a single T1929A synonymous nucleotide substitution was observed. This observation is significant, because this substitution has the potential to alter probe-amplicon hybridization necessary for discrimination by allele-specific probes that cover this region. Based on the FKS1 sequence consensus data, four allele-specific molecular-beacon probes that covered nucleotides 1920 to 1944 were designed. One probe was complementary to the wild-type FKS1 allele found in caspofungin-susceptible C. albicans strains, while three probes were complementary to mutant FKS1 alleles (C1934A, C1934T, T1933C) observed in caspofungin-resistant isolates (Table 1). All the molecular-beacon probes had identical 6-nucleotide stem domains, 5′ CGCGAG and CTCGCG 3′, and were synthesized with a wobble base consisting of A and T at a 50:50 ratio at the FKS1 single-nucleotide polymorphism (SNP) (position 1929) to ensure their hybridization to target sequences. Hybridization profiles were determined for molecular-beacon probes against eight DNA oligonucleotide templates representing the wild-type FKS1 and different FKS1 alleles bearing caspofungin resistance mutations at positions 1933 and 1934, as well as the SNP at position 1929 (Table 2). The Tm values for molecular beacons and their fully complementary DNA targets were close to each other within the temperature range 62.7 to 64.0°C. Such uniformity, which is essential to facilitate multiplex application of the probes in a single assay, was achieved by varying the length of the probe domain sequence for individual beacons. Molecular beacons FKS1(WT), FKS1(T1933C), FKS1(C1934A), and FKS1(C1934T) had probe domains of 24, 23, 25, and 25 nucleotides, respectively.
Isolation of spontaneous caspofungin-resistant C. albicans mutants.
Spontaneous mutants of C. albicans strains SC5314 and M70 resistant to caspofungin were isolated by direct selection on solid growth media containing 4 μg/ml (40 times the MIC) caspofungin. The frequency of formation of spontaneous caspofungin-resistant derivatives for both strains was <10−8 mutations per cell per generation. For both strains, the formation of rare shrunken, slow-growing colonies on the plates with caspofungin was observed. The colonies were transferred to fresh plates containing the same amount of caspofungin, and after prolonged incubation for >10 days, a small fraction of shrunken colonies gave rise to smooth, fast-growing derivatives that were able to propagate on caspofungin-containing media after reinoculation. In total, 35 and 50 isolates with confirmed resistance to caspofungin were isolated for strains SC5314 and M70, respectively. Microdilution-based testing of susceptibility to caspofungin revealed caspofungin MICs of >16 μg/ml for all laboratory-derived isolates. All caspofungin-resistant mutants demonstrated cross-resistance to micafungin and anidulafungin. Derivatives of strain M70 were highly resistant to micafungin (MIC at which 80% of isolates were inhibited [MIC80], ≥16 mg/ml) and anidulafungin (MIC80, ≥2 mg/ml), whereas SC5314 derivatives showed lower levels, with MIC80s of 1.0 to 2.0 μg/ml and 0.5 to 1 mg/ml for micafungin and anidulafungin, respectively.
Genotyping of caspofungin resistance mutations.
Real-time PCR with the allele-specific molecular beacons was used to identify fks1 mutant alleles in the mutant collections. Each chromosomal DNA sample was subjected to four separate reactions with individual molecular-beacon probes representing the different reduced-susceptibility alleles. Mutant FKS1 alleles were found for all 35 caspofungin-resistant derivatives of strain SC5314. A majority of them (n = 20) had the heterozygous (FKS1/fks1) mutation T1933C, while 15 mutants contained the homozygous (fks1/fks1) mutation T1933C, C1934A, or C1934T. In the case of heterozygosity at T1933C, two signals of similar magnitude from molecular beacons FKS1(WT) and FKS1(T1933C) were reported (Fig. 1). DNA samples with homozygous FKS1 mutations yielded distinct responses from corresponding mutant molecular beacons, with no signal from FKS1(WT). Conversely, chromosomal DNA from the parental strain SC5314 interacted only with the FKS1(WT) probe, while no fluorescence was detected from the mutant beacons. Genotyping of FKS1 alleles of caspofungin-resistant derivatives of strain M70 revealed the known mutation T1933C, C1934A, or C1934T in 44 out of 50 samples. As with the SC5314 mutants, a majority of M70 derivatives acquired heterozygous mutation T1933C. It was found that 25 of 50 strains with decreased susceptibility harbored the T1933C substitution in one FKS1 copy. Heterozygous mutations C1934A and C1934T were detected in six strains. Besides heterozygous mutations at positions 1933 and 1934 of FKS1, homozygous substitutions at these sites, identified by specific hybridization with the corresponding mutant molecular beacon FKS1(T1933C), FKS1(C1934A), or FKS1(C1934T), were also detected in 13 strains. All heterozygous and homozygous mutations detected in FKS1 by real-time PCR with molecular-beacon detection were confirmed by DNA sequencing (Table 3). DNA sequencing showed that all derivatives of strain SC5314 had the synonymous SNP T1929A, whereas derivatives of strain M70 lacked it, as expected.
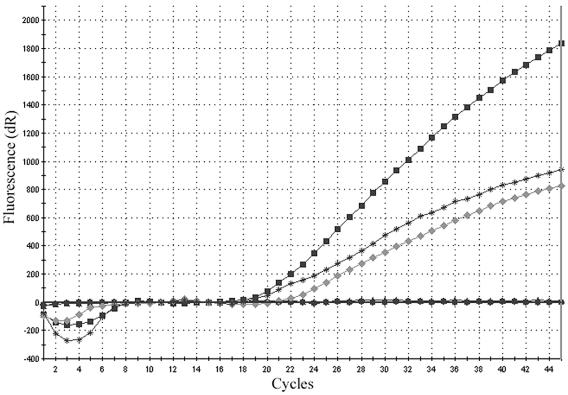
FIG. 1.
Detection of the T1933C mutation in both the homozygous and heterozygous states. The graph summarizes results of four separate PCRs with individual allele-specific molecular beacons and DNA targets. Squares, FKS1(T1933C) beacon plus DNA of the FKS1 allele with a homozygous T1933C mutation; circles, FKS1(WT) beacon plus DNA of the FKS1 allele with a homozygous T1933C mutation; asterisks, FKS1(T1933C) beacon plus DNA of the FKS1 allele with a heterozygous T1933C mutation; diamonds, FKS1(WT) beacon plus DNA of the FKS1 allele with a heterozygous T1933C mutation.
TABLE 3.
Distribution of FKS1 mutations among C. albicans caspofungin-resistant derivatives
| Mutation(s) in FKS1 | Mutation character | Amino acid change(s) | No. (%) of mutants derived from the following parental strain bearing the indicated mutationa:
|
|
|---|---|---|---|---|
| SC5314 | M70 | |||
| T1922C | Homozygous | F641S | 0 (0) | 5 (10) |
| T1933C | Heterozygous | S645P | 20 (57) | 25 (50) |
| T1933C | Homozygous | S645P | 5 (14) | 3 (6) |
| C1934A | Heterozygous | S645Y | 0 (0) | 3 (6) |
| C1934A | Homozygous | S645Y | 9 (26) | 7 (14) |
| C1934T | Heterozygous | S645F | 0 (0) | 3 (6) |
| C1934T | Homozygous | S645F | 1 (3) | 3 (6) |
| G1932T C1934G | Homozygous | L644F S645C | 0 (0) | 1 (2) |
Relative to a total of 35 mutants derived from strain SC5314 and a total of 50 mutants derived from strain M70.
Identifying new fks1 mutations.
In five strains, PCR amplification of chromosomal DNA was detected only weakly by the wild-type molecular beacon and not at all by the mutant molecular beacons. One strain was not detected by either the wild-type or the mutant molecular beacons. Given the allele specificity of the probes, these data suggested that the template sequence within the target region was altered in an unknown manner. Those six M70 derivatives were evaluated by direct DNA sequencing. The existence of a new homozygous mutation, T1922C, encoding an F641S substitution, was found in the FKS1 genes of five M70 derivatives. DNA sequencing of the sixth isolate, which reacted with neither wild-type nor mutant molecular-beacon probes, uncovered two new homozygous mutations, G1932T and C1934G, encoding the double substitution L644F and S645C.
Multiplex real-time detection of FKS1 mutations.
The application of allele-specific FKS1 molecular beacons made possible identification of known mutations in the C. albicans FKS1 gene. We further investigated the possibility of combining the FKS1 molecular beacons in a multiplex real-time PCR format suitable for simultaneous assessment of such mutations in a given DNA sample. The probe recognizing the wild-type sequence was labeled with FAM, while all mutant probes were labeled with HEX. Molecular beacons were pooled and used in a combined real-time PCR. Chromosomal DNAs from wild-type strains SC5314 and M70 and from 12 caspofungin-resistant derivatives of these strains representing 12 different FKS1 alleles listed in Table 3, were used as templates for multiplex real-time PCR. Caspofungin-susceptible and caspofungin-resistant phenotypes were identified by the nature of the fluorescence output. Only FAM fluorescence was observed when DNAs from susceptible strains SC5314 and M70 were subjected to multiplex real-time PCR. Only HEX fluorescence was reported in multiplex real-time PCR with DNAs bearing the homozygous mutation T1933C, C1934A, or C1934T in FKS1. Both FAM and HEX signals of equal magnitude were detected when the DNA analyzed was from strains known to have heterozygous mutation T1933C, C1934A, or C1934T in the FKS1 gene. Multiplex real-time PCR with chromosomal DNA from the strain with the two new mutations G1932T and C1934G in FKS1 yielded neither FAM nor HEX fluorescence. A minor FAM signal was observed in the reaction with chromosomal DNA from the strain containing the homozygous mutation T1922C, although this signal did not interfere with the final assessment of susceptibility. Overall, the multiplex panel performed as well as assays with individual molecular-beacon probes.
DISCUSSION
FKS1 as a prominent resistance mechanism.
Since echinocandins target glucan synthase, the machinery responsible for producing the major cell wall biopolymer, it was anticipated that cells exposed to the drug would induce changes at the target. It was recently reported that reduced susceptibility to caspofungin in clinical and laboratory strains of Candida albicans is associated with mutations in FKS1 at codon 645 in which serine is replaced by proline, tyrosine, or phenylalanine (2). These mutations permit growth of C. albicans in the presence of caspofungin at >16 μg/ml, increase the 50% inhibitory concentration of caspofungin for glucan synthase >500-fold, and shift by >100-fold the amount of the drug needed to reduce fungal burdens in target organs in murine infection models (2). The reduced-susceptibility phenotype was observed with both homozygous and heterozygous alleles present, indicating that expression of mutant FKS1 was dominant. To better understand the nature of FKS1 mutations in echinocandin resistance, we isolated two large collections of caspofungin-resistant isolates from two genetically distinct C. albicans strains. The generation of mutants with reduced susceptibility to caspofungin was a rare event and was consistent with a mutation frequency of <10−8 mutants per viable cell. A total of 85 spontaneous resistant mutants were analyzed by allele-specific molecular beacons representing wild-type and previously identified mutant sequences (Table 1). As observed with clinical isolates (2), most nucleotide substitutions were found in codon 645 of FKS1 (Table 3). Of the resistant mutants, 62% (53/85) displayed a characteristic S645P substitution expressed as either a homozygous or a heterozygous mutation in FKS1. Two other prominent amino acid substitutions at this locus, S645Y and S645F, were found at frequencies of 22% (19/85) and 8% (7/85), respectively. The allele specificity of the molecular-beacon sequencing probes helped to reveal three new mutations not observed previously in clinical isolates (2), T1922C, G1932T, and C1934G, encoding the F641S, L644F, and S645C substitutions, respectively (Table 3). The newly discovered mutations were seen only in the progeny of strain M70, with a relatively low frequency. There were no mutants recovered with mutations in the second region, defined by the Arg1361 locus of FKS1p (2), suggesting that this region contributes low-frequency resistance under selection pressure due to drug exposure.
The FKS1p-mediated mechanism of resistance examined in this study is not the only mode by which cells can modulate sensitivity to echinocandin drugs. The fungal cell wall is a dynamic structure, and resistance can potentially arise from genetic modulation of several cellular biosynthetic and regulatory pathways (10, 21). A complex network of pathways could account for slightly elevated MICs for some Candida isolates or for the observed in vitro high-dose paradox, in which cells appear to regain susceptibility at high levels of a drug (23). While these pathways have the potential to contribute to clinical resistance, it is important to distinguish these low-level drug tolerance and adaptive mechanisms from the FKS1p-mediated mechanisms that have been observed in clinical isolates and can result in treatment failure.
Molecular detection of mutations.
A wide range of molecular methods for mutation analysis and SNP genotyping are available (17). Among these, real-time PCR with detection by self-reporting molecular-beacon probes represents a powerful approach. Due to their hairpin structure, the thermodynamically conditioned equilibrium between intra- and intermolecular hybridization allows molecular beacons to distinguish closely related target sequences with higher specificity and within a wider temperature range than corresponding linear probes (3). The discriminatory power of molecular beacons has been successfully applied for analysis of mutations resulting in antibiotic resistance, allele differentiation, both homozygous and heterozygous SNPs, and a number of other applications (2, 11-13, 20, 22, 25).
The selectivity of molecular beacons was ideal for analysis of FKS1 alleles conferring reduced susceptibility to caspofungin. The assay focused on FKS1 codon 645 mutations T1933C, C1934A, and C1934T, which have been identified in clinical isolates of C. albicans showing reduced susceptibility to caspofungin. The design of allele-specific probes for detecting resistance alleles in the target region was complicated by the fact that a synonymous SNP, T1929A, was located within this domain. To accommodate this SNP, which appears in a significant proportion of clinical isolates, the probes were designed with a wobble base at the position corresponding to SNP. Introduction of the wobble base into the probe decreased the absolute fluorescence output, since at the given conditions only half of the probe molecular pool would bind specifically to the complementary target DNA. Nevertheless, the gain from the higher versatility of degenerate probes far exceeded the relative loss of fluorescence intensity, since such probes were reactive for the entire C. albicans population regardless of either the presence or the absence of the T1929A SNP in FKS1.
Overall, the application of individual molecular beacons allowed genotyping of all 85 caspofungin-resistant derivatives. The three mutant alleles targeted by molecular beacons were correctly identified in 79 strains. The application of probes recognizing both wild-type and mutant alleles facilitated the identification of three new mutations in six strains, which were fully elaborated by DNA sequence analysis. Multiplexing of all four molecular beacons in a single reaction produced a universal closed-tube assay for detection of caspofungin resistance mutations in FKS1. In the assay, the FAM fluorescent signal showed the presence of the wild-type FKS1 allele, whereas HEX fluorescence reported the presence of either the T1933C, C1934A, or C1934T FKS1 mutation in the DNA sample. DNA extracted from C. albicans strains heterozygous for any of these three mutations yielded both FAM and HEX signals. Thus, the assay allowed confident detection of known mutations in the C. albicans FKS1 gene conferring reduced susceptibility to caspofungin in both the homozygous and heterozygous states. The assay was also sensitive enough to detect the presence of other mutations in the region that can influence drug susceptibility. As molecular diagnostics continue to evolve, probes recognizing well-characterized resistance mechanisms have the potential to be used in routine diagnostic applications.
In conclusion, we have demonstrated that caspofungin resistance is linked at a high frequency to mutations in the FKS1 gene of C. albicans and most prominently to alterations of serine 645 of Fks1p. Furthermore, a real-time PCR assay utilizing allele-specific molecular beacons suitable for multiplex applications was employed for identification of these mutations. This tool can be expanded as new FKS1 mutations are identified, and it has value for high-throughput screening of clinical isolates for resistance by using characteristic FKS1 mutations as a surrogate marker.
Acknowledgments
This work was supported by NIH grant AI055767 to D.S.P.
REFERENCES
- 1.Bachmann, S. P., T. F. Patterson, and J. L. Lopez-Ribot. 2002. In vitro activity of caspofungin (MK-0991) against Candida albicans clinical isolates displaying different mechanisms of azole resistance. J. Clin. Microbiol. 40:2228-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balashov, S. V., R. Gardiner, S. Park, and D. S. Perlin. 2005. Rapid, high-throughput, multiplex, real-time PCR for identification of mutations in the cyp51A gene of Aspergillus fumigatus that confer resistance to itraconazole. J. Clin. Microbiol. 43:214-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonnet, G., S. Tyagi, A. Libchaber, and F. R. Kramer. 1999. Thermodynamic basis of the enhanced specificity of structured DNA probes. Proc. Natl. Acad. Sci. USA 96:6171-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cabello, M. A., G. Platas, J. Collado, M. T. Diez, I. Martin, F. Vicente, M. Meinz, J. C. Onishi, C. Douglas, J. Thompson, M. B. Kurtz, R. E. Schwartz, G. F. Bills, R. A. Giacobbe, G. K. Abruzzo, A. M. Flattery, L. Kong, and F. Pelaez. 2001. Arundifungin, a novel antifungal compound produced by fungi: biological activity and taxonomy of the producing organisms. Int. Microbiol. 4:93-102. [DOI] [PubMed] [Google Scholar]
- 5.Chandrasekar, P. H., and E. K. Manavathu. 2002. Caspofungin. Drugs Today (Barcelona) 38:829-846. [DOI] [PubMed] [Google Scholar]
- 6.Denning, D. W. 2002. Echinocandins: a new class of antifungal. J. Antimicrob. Chemother. 49:889-891. [DOI] [PubMed] [Google Scholar]
- 7.Diekema, D. J., S. A. Messer, R. J. Hollis, R. N. Jones, and M. A. Pfaller. 2003. Activities of caspofungin, itraconazole, posaconazole, ravuconazole, voriconazole, and amphotericin B against 448 recent clinical isolates of filamentous fungi. J. Clin. Microbiol. 41:3623-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones, T., N. A. Federspiel, H. Chibana, J. Dungan, S. Kalman, B. B. Magee, G. Newport, Y. R. Thorstenson, N. Agabian, P. T. Magee, R. W. Davis, and S. Scherer. 2004. The diploid genome sequence of Candida albicans. Proc. Natl. Acad. Sci. USA 101:7329-7334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurtz, M. B., and C. M. Douglas. 1997. Lipopeptide inhibitors of fungal glucan synthase. J. Med. Vet. Mycol. 35:79-86. [DOI] [PubMed] [Google Scholar]
- 10.Lesage, G., A. M. Sdicu, P. Menard, J. Shapiro, S. Hussein, and H. Bussey. 2004. Analysis of β-1,3-glucan assembly in Saccharomyces cerevisiae using a synthetic interaction network and altered sensitivity to caspofungin. Genetics 167:35-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin, S. Y., W. Probert, M. Lo, and E. Desmond. 2004. Rapid detection of isoniazid and rifampin resistance mutations in Mycobacterium tuberculosis complex from cultures or smear-positive sputa by use of molecular beacons. J. Clin. Microbiol. 42:4204-4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marras, S. A., F. R. Kramer, and S. Tyagi. 2003. Genotyping SNPs with molecular beacons. Methods Mol. Biol. 212:111-128. [DOI] [PubMed] [Google Scholar]
- 13.Marras, S. A., F. R. Kramer, and S. Tyagi. 1999. Multiplex detection of single-nucleotide variations using molecular beacons. Genet. Anal. 14:151-156. [DOI] [PubMed] [Google Scholar]
- 14.National Committee for Clinical Laboratory Standards. 2002. Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard, 2nd ed. NCCLS document M27-A2. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 15.Pacetti, S. A., and S. P. Gelone. 2003. Caspofungin acetate for treatment of invasive fungal infections. Ann. Pharmacother. 37:90-98. [DOI] [PubMed] [Google Scholar]
- 16.Park, S., R. Kelly, J. N. Kahn, J. Robles, M. J. Hsu, E. Register, W. Li, V. Vyas, H. Fan, G. Abruzzo, A. Flattery, C. Gill, G. Chrebet, S. A. Parent, M. Kurtz, H. Teppler, C. M. Douglas, and D. S. Perlin. 2005. Specific substitutions in the echinocandin target Fks1p account for reduced susceptibility of rare laboratory and clinical Candida sp. isolates. Antimicrob. Agents Chemother. 49:3264-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perlin, D. S., and S. Park. 2001. Rapid identification of fungal pathogens: molecular approaches for a new millennium. Rev. Med. Microbiol. 12(Suppl.):S13-S20. [Google Scholar]
- 18.Pfaller, M. A., D. J. Diekema, S. A. Messer, R. J. Hollis, and R. N. Jones. 2003. In vitro activities of caspofungin compared with those of fluconazole and itraconazole against 3,959 clinical isolates of Candida spp., including 157 fluconazole-resistant isolates. Antimicrob. Agents Chemother. 47:1068-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfaller, M. A., S. A. Messer, L. Boyken, C. Rice, S. Tendolkar, R. J. Hollis, and D. J. Diekema. 2003. Caspofungin activity against clinical isolates of fluconazole-resistant Candida. J. Clin. Microbiol. 41:5729-5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piatek, A. S., A. Telenti, M. R. Murray, H. El-Hajj, W. R. Jacobs, Jr., F. R. Kramer, and D. Alland. 2000. Genotypic analysis of Mycobacterium tuberculosis in two distinct populations using molecular beacons: implications for rapid susceptibility testing. Antimicrob. Agents Chemother. 44:103-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reinoso-Martin, C., C. Schuller, M. Schuetzer-Muehlbauer, and K. Kuchler. 2003. The yeast protein kinase C cell integrity pathway mediates tolerance to the antifungal drug caspofungin through activation of Slt2p mitogen-activated protein kinase signaling. Eukaryot. Cell 2:1200-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rice, J. E., J. A. Sanchez, K. E. Pierce, and L. J. Wangh. 2002. Real-time PCR with molecular beacons provides a highly accurate assay for detection of Tay-Sachs alleles in single cells. Prenat. Diagn. 22:1130-1134. [DOI] [PubMed] [Google Scholar]
- 23.Stevens, D. A., T. C. White, D. S. Perlin, and C. P. Selitrennikoff. 2005. Studies of the paradoxical effect of caspofungin at high drug concentrations. Diagn. Microbiol. Infect. Dis. 51:173-178. [DOI] [PubMed] [Google Scholar]
- 24.Stone, E. A., H. B. Fung, and H. L. Kirschenbaum. 2002. Caspofungin: an echinocandin antifungal agent. Clin. Ther. 24:351-377. (Discussion, 24:329). [DOI] [PubMed] [Google Scholar]
- 25.Templeton, K. E., S. A. Scheltinga, M. F. Beersma, A. C. Kroes, and E. C. Claas. 2004. Rapid and sensitive method using multiplex real-time PCR for diagnosis of infections by influenza A and influenza B viruses, respiratory syncytial virus, and parainfluenza viruses 1, 2, 3, and 4. J. Clin. Microbiol. 42:1564-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tyagi, S., D. P. Bratu, and F. R. Kramer. 1998. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 16:49-53. [DOI] [PubMed] [Google Scholar]