Figure 1.
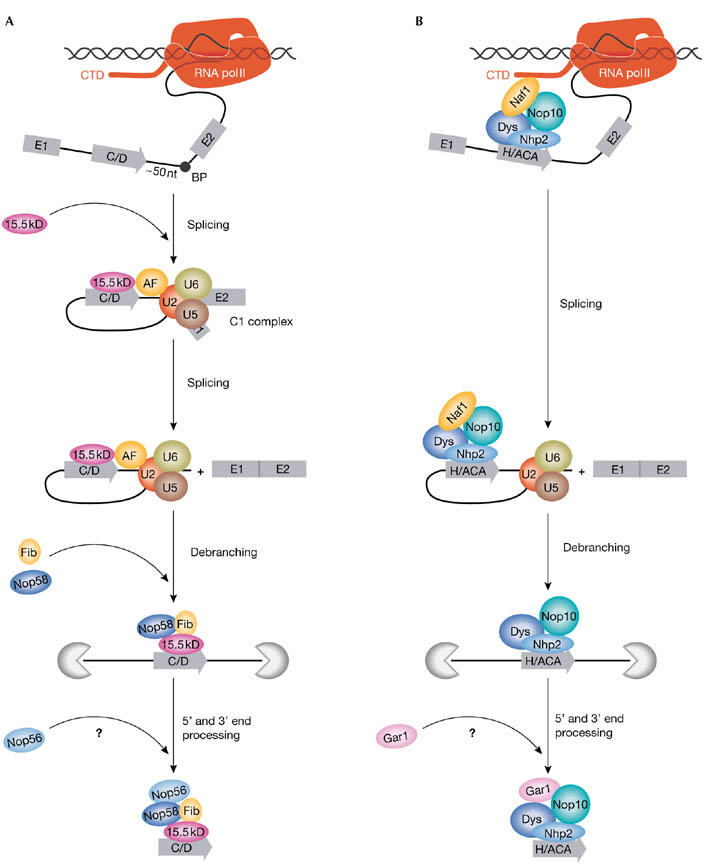
Models for coupling intronic small-nucleolar ribonucleoprotein assembly with pre-messenger RNA transcription and splicing. (A) Splicingdependent assembly of intronic box C/D small-nucleolar ribonucleoproteins (snoRNPs). The majority of mammalian intronic box C/D snoRNAs are located about 50 nucleotides (nt) upstream of the branch point (BP) and are processed in a splicing-dependent manner. At the C1-complex stage of splicing, the 15.5 kDa (kD) central core protein is actively recruited to the intronic snoRNA by a putative assembly factor (AF) that probably interacts with splicing factors associated with the BP region. Fibrillarin (Fib) and the nucleolar protein Nop58 are believed to bind directly after 15.5 kDa docking, but Nop56 might bind at a later stage of snoRNP biogenesis. (B) Transcription-dependent assembly of intronic box H/ACA snoRNPs.Assembly of H/ACA presnoRNPs occurs co-transcriptionally. Nuclear-assembly factor 1 (Naf1), which is an essential H/ACA assembly factor, might interact with the carboxyterminal domain (CTD) of RNA polymerase (pol) II, and could facilitate recruitment and binding of the H/ACA core proteins to the nascent snoRNA sequences. The correct timing of glycine arginine rich 1 (Gar1) binding is still unclear. It is important to note that assembly of a minority group of box C/D snoRNPs that have suboptimal intronic positions and carry external intronic stem structures might follow a transcription-dependent processing pathway similar to that described for H/ACA snoRNPs (Hirose et al, 2003). Dys, dyskerin; Nhp2, non-histone chromosome protein 2.
