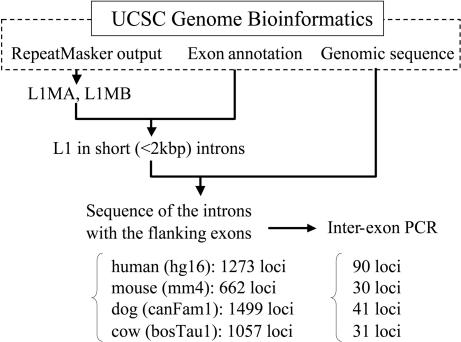
Fig. 1.
The in silico screening of genomic data for L1 sequences in introns performed in this study by using the databases available from University of California Santa Cruz Genome Bioinformatics (http://genome.ucsc.edu). The number of L1 insertions found and the number of loci used in interexonic PCR are shown for each species. The L1MA and L1MB are L1 subfamilies used for insertion comparison in this study. hg16, mm4, canFam1, and bosTau1 denote the database versions of human, mouse, dog, and cow, respectively.