Abstract
The usefulness of digital clinical information is limited by difficulty in accessing that information. Information in electronic medical records (EMR) must be entered and stored at the appropriate level of granularity for individual patient care. However, benefits such as outcomes research and decision support require aggregation to clinical data -- "heart disease" as opposed to "S/P MI 1997" for example. The hierarchical relationships in an external reference terminology, such as SNOMED, can facilitate aggregation. This study examines whether by leveraging the knowledge built into SNOMED’s hierarchical structure, one can simplify the query process without degrading the query results.
Methodology
Concepts from the EMR in the cardiovascular domain were mapped to SNOMED concepts. Next, actual query requests of the data center were reviewed for concepts that had been mapped to SNOMED. The data center then used its standard methods to return a set of de-identified patient keys for each concept. Sets of patient keys were then also obtained using SNOMED based queries written in Oracle 8i SQL.
Results.
The precision of each query exceeded 0.97 with the exception of Beta Blocker which was 0.95. The recall of the SNOMED -based queries for each concept is shown in Table 1. In 14 of 16 queries the recall exceeded 0.90. For CAD, its failure to meet 0.90 recall was due to both a failure to map the local vocabulary concept ‘atherosclerotic heart disease’ to SNOMED and the data center’s ability to include subjects who had had a procedure that indicated the presence of CAD, such as Coronary Artery Bypass Graft (CABG). Type I diabetes did not reach 0.90 recall solely because the data center included the concept of ‘insulin dependant diabetes mellitus’ while this concept was not under the type I diabetes hierarchy in SNOMED.
Table 1.
Recall of SNOMED -based queries
| Concept | Subjects | Recall |
|---|---|---|
| Myocardial Infarction | 2581 | 0.979 |
| Coronary Artery Disease (CAD) | 17414 | 0.893 |
| Heart Failure | 6997 | 0.921 |
| Hypertension | 99559 | 0.979 |
| Type I Diabetes Mellitus | 3323 | 0.741 |
| Type II Diabetes Mellitus | 27899 | 0.987 |
| Hyperlipidemia | 69507 | 1.000 |
| HMGCoA Reductase Inhibitor | 58648 | 0.990 |
| Niacin | 1489 | 0.952 |
| Insulin | 11734 | 0.979 |
| Biguanide (Metformin) | 18265 | 0.944 |
| Aspirin | 61475 | 0.960 |
| Thiazide Diuretic | 59151 | 0.989 |
| Beta Blocker | 55358 | 0.993 |
| ACE Inhibitor | 61326 | 0.989 |
| Angiotensin II Receptor Blocker | 19619 | 0.989 |
Discussion.
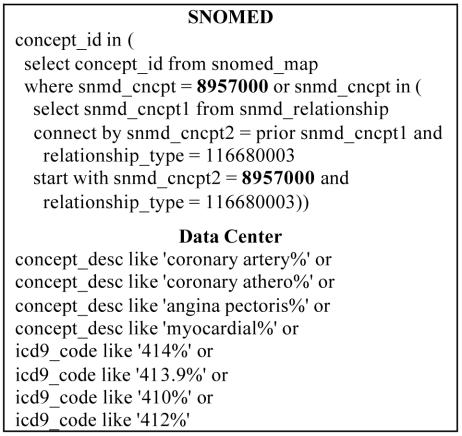
Figure 1 demonstrates how SNOMED simplifies the query process. To look for a different concept using SNOMED, one only has to substitute the new SNOMED concept id in place of ‘8957000’. Without using SNOMED, one would need to construct the new query using knowledge of clinical medicine, coding schemes, and the database structure. This study has demonstrated that it is possible to apply SNOMED to a highly granular clinical terminology and simplify the query process, thereby making it less costly to develop and maintain queries without significant degradation of query results.
Figure 1.
CAD SQL search criteria