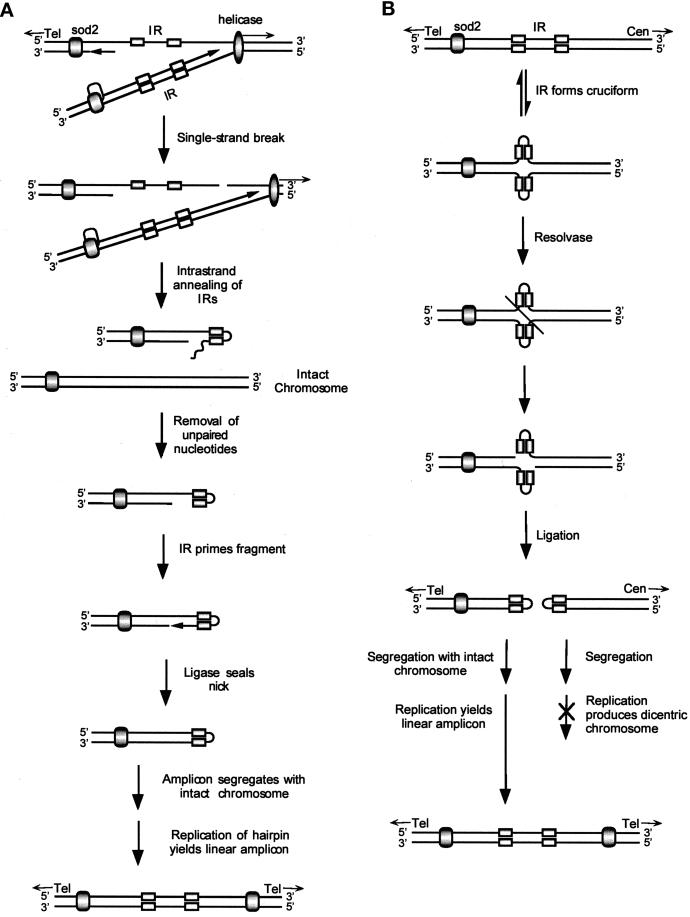
Figure 8.
Replication and recombination models for the 225-kb amplicon. (A) Replication model. sod2 is flanked by a telomere (Tel) on the left and an inverted repeat (IR) on the right. sod2 is shown as a shaded oval, the inverted repeats are shown as rectangles, and the helicase/replication complex is shown as an elongated oval. A single-strand DNA break on the lagging strand, to the right of the inverted repeat and ahead of the replication fork, allows the inverted repeat to form a hairpin. Replication of the other DNA strand is unaffected. Removal of unpaired nucleotides allows the hairpin to prime DNA synthesis. After ligase seals the nick, a larger hairpin, consisting of a duplication of the region from the inverted repeat to the telomere, is formed. The large hairpin and intact chromosome I must segregate together to yield a viable cell resistant to LiCl. The mature palindrome is formed in the next cell cycle. (B) Recombination model. The inverted repeat to the right of sod2 forms a cruciform analogous to a Holliday structure. If resolvase cuts across the axis of symmetry and ligase joins the free ends of the different DNA strands, two hairpins will be produced that include sod2 and the centromere (Cen), respectively. The sod2 hairpin must segregate with intact chromosome I to form a viable cell resistant to LiCl. The next round of replication produces the mature palindrome. Because the fission yeast S. pombe is haploid during selection with LiCl, this mitotic recombination event must occur after replication of the DNA.