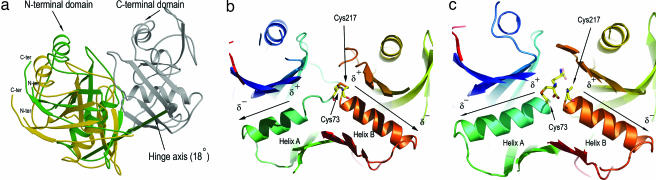
Fig. 2.
Conformational changes on inhibitor binding. (a) Shell-like domain closure upon binding AziDAP inhibitors. The hinge rotation axis (≈18°), represented by an arrow, was determined by superimposition of the C-terminal domain of the native, inactive DAP epimerase (PDB ID code 1GQZ) with the C-terminal domain of the ll-AziDAP-inhibited DAP epimerase (rmsd = 0.32 Å for 145 Cα atoms). The N-terminal domain of the unbound enzyme is shown in gold, and that of the inhibited DAP epimerase is shown in green. The arrow coloring indicates the direction of rotation of the N-terminal domain (green) relative to the C-terminal domain (gray). (b) Environments of the two opposed α-helices in the native unbound DAP epimerase (PDB ID code 1GQZ). The N-terminal four-helix A residues, Gly-74 to Ala-77, are partially unwound in this structure. Helix A is colored teal; helix B (Gly-218 to Met-230) in the C-terminal domain of DAP epimerase is fully formed and colored orange. The two catalytic cysteines (Cys-73 and Cys-217) are involved in a disulfide link. (c) Helices A and B, in the presence of the bound inhibitor AziDAP, covalently linked to Sγ of Cys-73. The carbon atoms of the inhibitor are colored yellow (nitrogen atoms, blue; oxygen, red). Helix A (Gly-74 to Leu-86) is on the left, and helix B is on the right. The opposing dipole moments are directed at the α-carbon carboxylate.