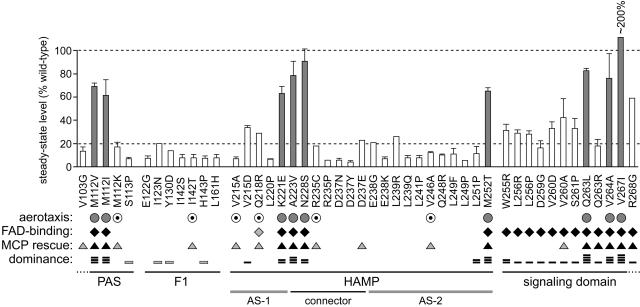
FIG. 5.
Summary of Aer mutant (Aer*) properties. The residue numbers and amino acid replacements of pMB1-Aer* characterized in this study are listed below the histogram, which shows the steady-state expression level of each mutant protein, determined as detailed in Materials and Methods. Shaded vertical bars indicate Aer proteins with CW-biased signal output, as determined by flagellar rotation pattern. Symbols: for aerotaxis (UU1250 colony phenotype on tryptone soft agar), shaded circles, fuzzy, and dotted circles, leaky; for FAD binding (ability to increase cellular FAD levels when highly induced), black diamonds, increases comparable to those produced by wild-type Aer, and shaded diamonds, intermediate-level increase; for MCP rescue (recovery of aerotactic ability in UU1117), black triangles, fully rescued, and shaded triangles, partial rescue or rescue at high induction; for dominance (ability to block aerotaxis in UU1250 carrying a wild-type aer plasmid), one bar reduced colony size on soft agar (shading denotes effects seen only at high induction); two bars, impaired ring formation; three bars, reduced size and impaired ring formation; and four bars, null phenotype.