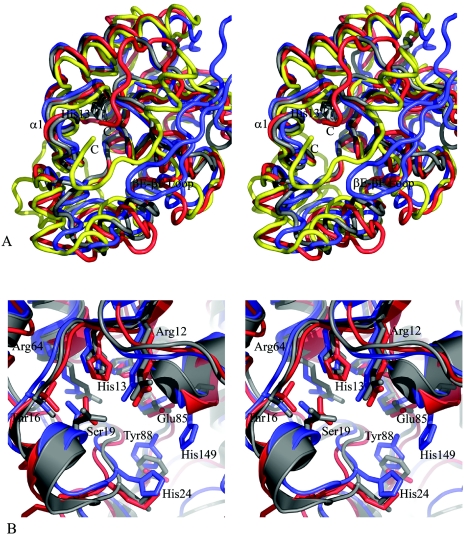
FIG. 4.
Comparison of Rv3214 structure with homologous proteins. (A) Stereoview showing a superposition of the Rv3214 structure (blue) onto those of PhoE (red), E. coli dPGM (gold), and the T. thermophilus dPGM homolog (gray). The view is looking down into the active site, with the essential histidine (His13 in Rv3214) shown in dark gray at the bottom. Differences in Rv3214 result from inward movement of several flanking helices and from the presence of the βE-βF loop (labeled) from the other monomer of the dimer; this helps enclose the active site, a role filled by the C-terminal regions of dPGM and PhoE. The T. thermophilus protein has no equivalent feature and a much more open active site. (B) Stereoview of a superposition of the active sites of Rv3214 (blue), PhoE (red), and the T. thermophilus enzyme (gray). Residue labels are for Rv3214.