Abstract
Magnetospirillum magnetotacticum possesses intracellular magnetite particles with a chain-like structure, termed magnetosomes. The bacterium expresses 22-kDa and 12-kDa magnetosome-associated proteins, termed Mam22 (MamA) and Mam12 (MamC), respectively. In this study, we investigated the structure of the purified magnetosomes with transmission electron microscopic techniques and found that the magnetosomes consisted of four compartments, i.e., magnetite crystal, magnetosomal membrane, interparticle connection, and magnetosomal matrix. Furthermore, we determined the precise localizations of Mam22 and Mam12 using immunogold staining of the purified magnetosomes and ultrathin sections of the bacterial cells. Interestingly, most Mam22 existed in the magnetosomal matrix, whereas Mam12 was strictly localized in the magnetosomal membrane. Moreover, the recombinant Mam22 was attached to the magnetosomal matrix of the Mam22-deficient magnetosomes prepared by alkaline treatment, such as 0.1 M Caps-NaOH buffer (pH 11.0). The spatial localization of the magnetosome-associated proteins in the magnetosomal chain provides useful information to elucidate the functional roles of these proteins.
Magnetic bacteria are a phylogenetically diverse group of highly motile aquatic eubacteria that can orient in geomagnetic fields and that possibly search for microaerophillic environments using intracellular magnetite particles with a chain-like structure, termed magnetosomes (3). Many researchers have attempted to elucidate the synthesis processes of the intriguing magnetic particles since Blakemore et al. first isolated one species of the group, Magnetospirillum magnetotacticum MS-1, that was later used as a laboratory model (4).
Magnetosomes appear to be surrounded by a magnetosomal membrane and/or an organic matrix (2, 6, 12, 15, 23). Balkwill et al. reported that an individual magnetite crystal was enveloped by the magnetosomal membrane with a laminate structure (2). Gorby et al. reported an organic matrix surrounding the magnetosomes in M. magnetotacticum (6). The biochemical compositions of the magnetosomes have also been analyzed (6, 7). Gorby et al. showed that the magnetosomal membrane is composed of neutral lipids, free fatty acids, glycolipids, sulfolipids, and phospholipids (6). The magnetosome also contains various kinds of specific associated proteins, such as the tetratricopeptide repeat (TPR) protein, ion transporters, proteases, heme binding proteins, iron binding proteins, and hypothetical proteins, which are predicted from their deduced amino acid sequences (1, 6, 7, 13, 14, 16, 17). Recently, genes encoding magnetosome-associated proteins were identified within a putative gene island (the mam gene island) in the genomes of Magnetospirillum gryphiswaldense, M. magnetotacticum, and Magnetococcus sp. strain MC-1 (7, 8, 22, 24). The spontaneous deletion mutant of the mam island lacked any structures resembling magnetite crystals or membrane vesicles, suggesting that these magnetosome-associated proteins could be presumed to play major roles in the construction of magnetosomes (22, 24). The mechanisms of magnetosome synthesis in magnetic bacteria, however, remain elusive, because of a lack of information about the functions of these proteins.
Mam22, an abundant protein in the magnetosome, contains five TPR motifs and one putative TPR motif (16, 17). TPR motifs, which are widely found in the proteins of many kinds of organisms, consist of 3 to 16 tandem repeats of 34 amino acid residues and mediate protein-protein interactions to assemble the multiprotein complexes (5). Therefore, Mam22 of M. magnetotacticum may function as a receptor for the protein-protein interaction in magnetosomes, whereas Mam12 is a putative membrane protein with two transmembrane helices and showed no homology to known proteins. The genes of the two heterogeneous proteins, Mam22 and Mam12, are encoded in the same mam gene island as orthologous genes of mamA and mamC, respectively (22). Despite detailed analyses of these proteins at the genetic and molecular levels, it is necessary to know the physically precise locations of the magnetosome-associated proteins for a detailed explanation of the roles of these proteins in the magnetosome.
In this study, we examined the precise localizations of two magnetosome-associated proteins, Mam22 and Mam12, in the magnetosomes. We revealed that most Mam22 exists in the magnetosomal matrix of the magnetosome, while Mam12 was strictly localized on the magnetosomal membrane. Furthermore, we prepared the Mam22-deficient magnetosomes by treatment with 0.1 M Caps-NaOH buffer (pH 11.0) and found that the recombinant His-tagged Mam22 was specifically bound to the matrix of the alkaline-treated magnetosomes.
MATERIALS AND METHODS
Microorganisms and cultures.
M. magnetotacticum MS-1 (ATCC 31632) was cultured in a chemically defined liquid medium (MS-1 medium) under an O2 (1%)-N2 (99%) atmosphere at 25°C in the dark (4). The cells were harvested in early stationary phase using continuous centrifugation at 10,000 × g at 4°C. Escherichia coli strain BL21(DE3) (Novagen) containing pET15b-mam22, which was constructed by Okuda and Fukumori (16), was used for overproduction of His-tagged Mam22. E. coli was grown at 37°C under aerobic conditions in Luria-Bertani medium (19) with 50 μg/ml (final concentration) of ampicillin until the optical density at 600 nm reached 0.8 to 1.0, and then 1 mM isopropyl-β-d-thiogalactopyranoside (final concentration) was added to the culture for the induction of His-tagged Mam22. The cells were harvested using centrifugation at 8,000 × g for 15 min after 2 hours of incubation under the same conditions.
Physical and chemical measurements.
The protein contents were determined by the bicinchoninic acid method (BCA Protein Assay Kit; Pierce) with bovine serum albumin as a standard. Spectrophotometric measurements were carried out with a Shimadzu spectrophotometer (MPS-2000), using a 1-cm light path cuvette at 25°C. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed by the methods of Laemmli or Schägger and von Jagow (11, 20). For SDS-PAGE analyses, the proteins or magnetosomes were treated with SDS-PAGE sample buffer (2% SDS, 2% β-mercaptoethanol, and 50 mM Tris-HCl buffer [pH 6.8]) at 100°C for 3 min. The protein bands were stained with Coomassie brilliant blue G-250.
Magnetosome purification.
The frozen cells (ca. 25 g [wet weight]) were thawed out and suspended in 100 ml of 10 mM Tris-HCl buffer (pH 8.0) and disrupted with an ultrasonic oscillator, Branson model 450 (20 kHz; 80 W), for 10 min. After the suspension was centrifuged at 8,000 × g for 15 min, the pellet obtained was suspended in 100 ml of 10 mM Tris-HCl buffer (pH 8.0) and treated with an ultrasonic oscillator as described above. The suspension was centrifuged at 8,000 × g for 15 min, and the pellet obtained was suspended in 100 ml of 10 mM Tris-HCl buffer (pH 8.0). The suspension in a beaker was placed on a bar magnet (2.5 cm × 12 cm) for 1 h, and then the nonmagnetic fluid was removed by aspiration. The magnetosomes attracted to the magnet were carefully suspended in 100 ml of 10 mM Tris-HCl buffer (pH 8.0). This procedure with the magnet was repeated at least 10 times, and the purified magnetosomes were finally collected by centrifugation at 8,000 × g for 15 min. The precipitate was suspended in 10 mM Tris-HCl buffer (pH 8.0) and stored at −80°C. All purification steps were conducted at 4°C.
Selective solubilization of Mam22.
The aliquots of the purified magnetosomes (1 mg [wet weight]) were suspended in 10 μl (each) of 0.1 M glycine-HCl buffer (pH 1.8), 0.1 M glycine-HCl buffer (pH 2.5), 0.1 M Caps-NaOH buffer (pH 11.0), 0.1 M sodium borate buffer (pH 11.0), 3 M thiocyanate (pH 11.0), 3.5 M MgCl2, 5 M NaCl, 8 M urea, 5 M guanidine-HCl, and 10% dioxane. After the samples were incubated at 4°C for 16 h, the suspensions were centrifuged at 8,000 × g for 10 min. To investigate the efficiencies of selective solubilization of Mam22, the protein compositions of the resultant supernatants were analyzed by SDS-PAGE.
To prepare alkaline-treated magnetosomes, the purified magnetosomes (4 mg [wet weight]) were suspended in 50 μl of 0.1 M Caps-NaOH buffer (pH 11.0), and incubated at 4°C for 1 h. The magnetosome suspension was centrifuged at 8,000 × g for 10 min, and the pellet obtained was suspended in the same buffer. This step was repeated three times. After the resultant pellet was suspended in 1 ml of 10 mM Tris-HCl buffer (pH 8.0), the suspension stood for 30 min on a bar magnet (2.5 cm × 12 cm). The supernatant nonmagnetic fraction was removed by aspiration, and then the magnetic fraction was resuspended in 10 mM Tris-HCl (pH 8.0). The magnetosomes were washed with the same buffer at least three times and then used for immunogold staining and transmission electron microscopy (TEM) observation.
Reconstruction with recombinant Mam22.
For reconstruction with recombinant Mam22, the alkaline-treated magnetosomes (4 mg [wet weight]) were incubated with the purified His-tagged Mam22 (20 μg) in 50 μl of 10 mM Tris-HCl (pH 8.0) at 25°C for 16 h and then centrifuged at 8,000 × g for 10 min. The pellets obtained were washed with 1 ml of 10 mM Tris-HCl buffer (pH 8.0) and collected within 30 min by a magnet. The supernatant containing the unbound His-tagged Mam22 protein was removed by aspiration, and the magnetic fraction was resuspended in 10 mM Tris-HCl buffer (pH 8.0). This washing step was repeated at least six times.
Generation of polyclonal antibodies.
Polyclonal antibodies against Mam22 and Mam12 were generated by surgically implanting purified antigens in female New Zealand White rabbits (9). The His-tagged Mam22 was overexpressed in E. coli and purified by a method described previously (16). The purified His-tagged Mam22 was treated with a Thrombin kit (Novagen) at 20°C for 16 h to remove the His tag. The sample was resolved on SDS-PAGE, and the protein bands of Mam22 were excised from the gel. After the excised bands were homogenized with a minimal volume of 10 mM Tris-HCl buffer (pH 8.0) containing 0.1% SDS, the homogenate was incubated at room temperature for 16 h and centrifuged at 17,500 × g for 10 min. The Mam22 in the supernatant was concentrated by acetone precipitation and used as the antigen for generation of anti-Mam22 antibodies.
To prepare the antigen for generation of anti-Mam12 serum, the purified magnetosomes were treated with SDS-PAGE sample buffer (2% SDS, 2% β-mercaptoethanol, and 50 mM Tris-HCl [pH 6.8]) at 100°C for 3 min and centrifuged at 17,500 × g for 10 min. The supernatant obtained was resolved on SDS-PAGE, and the protein bands of Mam12 were excised and homogenized with a minimal volume of 10 mM Tris-HCl buffer (pH 8.0) containing 0.1% SDS. The homogenate was incubated at room temperature for 16 h and centrifuged at 17,500 × g for 10 min. The Mam12 in the supernatant was concentrated by acetone precipitation and used as the antigen for generation of anti-Mam12 antibodies.
The antigens (100 μg) were dissolved in phosphate-buffered saline (PBS) containing 0.1% SDS, emulsified, and injected into rabbits. The booster immunizations were delivered approximately every 2 weeks. Both sera were harvested after five booster immunizations and stored at −80°C with 0.02% sodium azide for preservation. The antibody activities were checked using an immunodiffusion method (18). According to immunoblotting analyses using the generated antibodies, the apparent molecular masses corresponding to Mam22 and Mam12 were recognized as single positive bands from solubilized magnetosome-associated proteins (data not shown).
Immunoblotting analyses.
The proteins separated by SDS-PAGE were transferred to polyvinylidene fluoride membranes (Hybond P; Amersham Bioscience) using an electroblotting method at 1 mA/cm2 for 2 h. Immunoreactivity species of anti-Mam22 and anti-Mam12 antibodies were detected at a dilution of 1:50,000. Goat anti-rabbit IgG conjugated to horseradish peroxidase (Amersham Bioscience) was used at a dilution of 1:10,000 for ECL Plus Western blotting detecting reagents (Amersham Bioscience). The chemifluoresence data were collected using a luminescent image analyzer, LAS 3000 (Fujifilm). The band intensities were quantified using Multi Gauge version 2.2 software (Fujifilm).
Specimen preparation for TEM.
To prepare the specimen for TEM observation, Formvar- and carbon-coated grids were put on a drop of the purified magnetosome suspension for about 1 min. Some of the grids were negatively stained with 4% uranyl acetate or 2% sodium tungstate for several seconds. The specimens were studied with a JEOL JEM 2000EX TEM operating at 120 kV in bright-field mode.
Immunogold labeling of Mam22 and Mam12 in the purified magnetosomes.
The grids with the purified magnetosomes were floated on a drop of water for 10 min and then incubated with 1% bovine serum albumin (BSA) in PBS for 5 min to block the nonspecific protein binding, followed by incubation with anti-Mam22 or anti-Mam12 rabbit polyclonal antibodies for 6 h at room temperature. The antisera were diluted 1:50 with 0.5% BSA in PBS. After rinses with 0.5% BSA in PBS three times for 1 min each time, the specimens were incubated with 5-nm- or 15-nm-diameter gold-conjugated goat anti-rabbit IgG (EY laboratories, Inc.) for 8 h at room temperature. After being rinsed three times with PBS for 1 min each time, the specimens were rinsed with deionized water twice for 3 min each time. As a cytochemical control, the primary antibody was replaced with preimmune serum. Moreover, after being immunostained, some of the specimens were negatively stained with 4% uranyl acetate or 2% sodium tungstate for several seconds. The specimens were observed by TEM under the same experimental conditions described above. To further clarify the localizations of Mam22 and Mam12 in the magnetosomal chains, the distances from the surfaces of the gold particles to the surfaces of the 1,000 magnetite particles were measured on TEM micrographs.
Immunogold labeling of Mam22 and Mam12 using ultrathin sections.
The bacterial cells at early stationary phase were concentrated by centrifugation and embedded in a low-melting-temperature agar (Agar Noble; Becton Dickinson), followed by cutting them into blocks smaller than 3 mm3. The blocks were fixed for 4 h at 4°C with PLP fixation solution (0.01 M NaIO4−, 0.075 M lysine hydrochloride, 0.0375 M phosphate buffer, 2.0% paraformaldehyde, pH 6.2), followed by two washes for 30 min each in a 0.1 M phosphate buffer (pH 7.3). The blocks were partially dehydrated through a graded ethanol series (30, 50, and 70% ethanol for 10 min each) and transferred into London Resin White from the 70% ethanol solution. After infiltration for 8 h, the blocks were embedded in gelatin capsules, which were polymerized at 50°C for 40 h. Ultrathin sections 60 nm thick were prepared using a microtome (Leica Ultracut R) equipped with a Diatome diamond knife and mounted on 200-mesh Formvar- and carbon-coated Ni grids for immunogold labeling.
The grids with ultrathin sections were floated on a drop of water for 20 min and then prepared by the same procedure with immunogold labeling of the purified magnetosomes described above. After the immunogold labeling, the ultrathin sections were stained with 4% uranyl acetate for 5 min and 0.4% lead citrate for 3 min. The specimens were observed by TEM under the same experimental conditions described above.
RESULTS
Fine structure of magnetosomes.
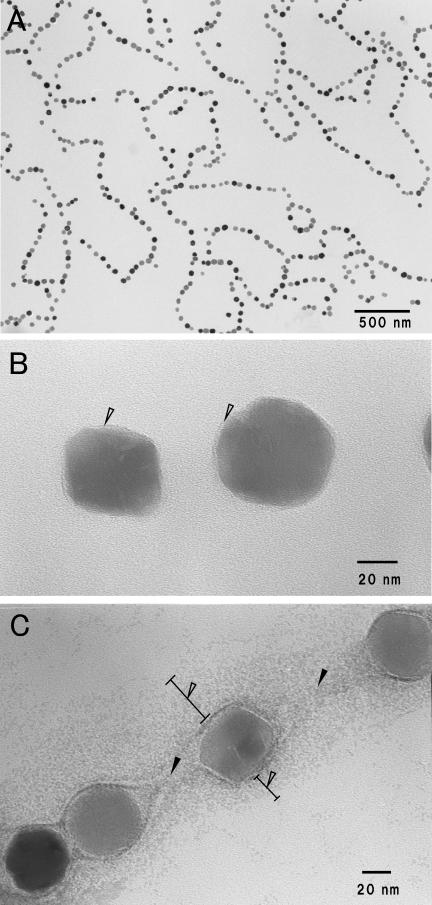
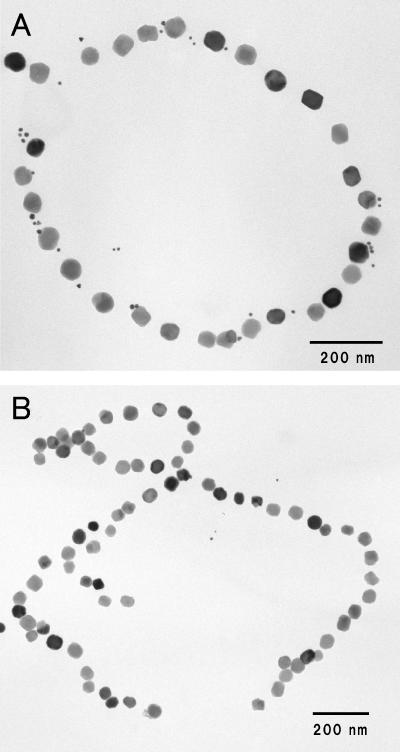
Figure 1A shows a transmission electron micrograph of the magnetosome preparations purified by the method described in Materials and Methods. Magnetite particles with diameters of 50.1 ± 6.2 nm were arranged in a linear array, the same as within the Magnetospirillum magnetotacticum MS-1 cells. TEM observation at a high magnification showed that the magnetosome individually had an electron-transparent layer which was visible on only part of the surface (Fig. 1B). The layer, which was 1.7 ± 0.5 nm thick on average, coincided with an electron-transparent layer of trilaminate magnetosomal membrane, as Gorby et al. previously reported (6). Furthermore, the magnetosomes with uranyl acetate staining revealed that there was an interparticle connection with a fibrous texture between the magnetite particles, parallel to a line of the magnetite particles, and it appeared to be elastic in the same direction as the line of magnetite particles (Fig. 1C). The texture is possibly caused by elongation of parts of the magnetosomal chain during the purification process or sample preparation for TEM observation. The other constituent of the magnetosomal matrix was also defined as a positively stained material that spread around a line of magnetite particles 108 ± 20 nm in width (Fig. 1C). The width was greater than those of the magnetosomal matrices in other magnetic bacterial cells previously reported by Taylor and Barry (23).
FIG.1.
Transmission electron micrographs of purified magnetosomes from M. magnetotacticum. (A) Low-magnification electron micrograph and (B) high-magnification micrograph of magnetosomes. Magnetite particles 50.1 ± 6.2 nm in diameter are surrounded by the electron-transparent layer (arrowheads). (C) The purified magnetosomes negatively stained with 4% uranyl acetate. Note that the magnetosomes are connected by electron-dense fibrous structures and interparticle connections (solid arrowheads) and surround an electron-dense region, the magnetosomal matrix (open arrowheads).
Selective solubilization of Mam22 from purified magnetosomes.
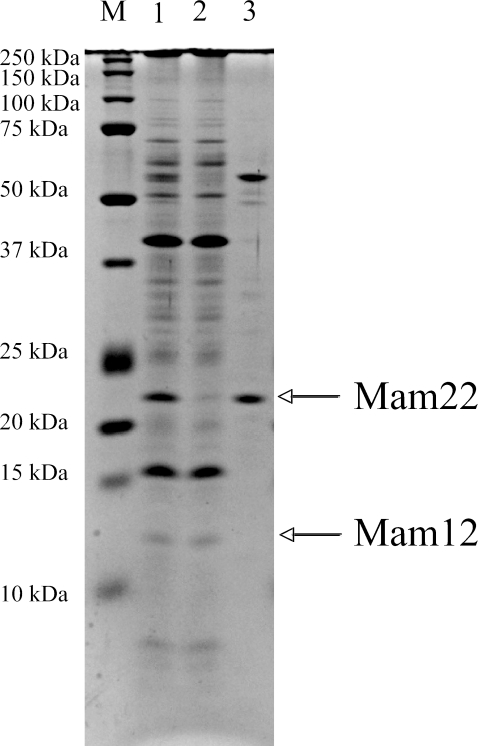
The binding characteristics of Mam22 and Mam12 in purified magnetosomes were examined by treatments of some solubilizing agents, such as acidic buffers (0.1 M glycine-HCl buffer, pH 1.8, and 0.1 M glycine-HCl buffer, pH 2.5), alkaline buffers (0.1 M CAPS-NaOH buffer, pH 11.0, and 0.1 M sodium borate, pH 11.0), chaotropic agent (3 M thiocyanate, pH 11.0), high-salt solutions (3.5 M MgCl2 and 5 M NaCl), dissociating agents (8 M urea and 5 M guanidine-HCl), and organic solvent (10% dioxane). The acidic buffers, high-salt solutions, and organic solvent had no effect on the solubilization of Mam22 (data not shown), while the alkaline buffers, chaotropic agent, and dissociating agents solubilized Mam22. In particular, 0.1 M Caps-NaOH buffer (pH 11.0) most effectively solubilized Mam22, but not Mam12 (Fig. 2). A 57-kDa protein was also solubilized by the alkaline treatment (Fig. 2) and was identified as cytochrome cd1-type nitrite reductase (25) using N-terminal amino acid sequence analysis. According to immunoblot analysis using anti-Mam22 antibodies, ca. 90% of the Mam22 protein was removed from the magnetosomes by three alkaline treatments (data not shown). These results suggest that Mam22 is loosely bound to the magnetosomes compared with other magnetosome proteins, including Mam12. In the rest of the study, we used 0.1 M Caps-NaOH buffer (pH 11.0) as the solubilizing agent for Mam22.
FIG. 2.
SDS-PAGE analyses of the proteins extracted from intact magnetosomes and alkaline-treated magnetosomes. Lane 1, SDS-PAGE analysis of the proteins extracted from the intact magnetosomes; lane 2, SDS-PAGE analysis of the proteins extracted from the magnetosomes that had been treated with 0.1 M Caps-NaOH (pH 11.0); lane 3, SDS-PAGE analysis of the proteins solubilized from magnetosomes by alkaline treatment with 0.1 M Caps-NaOH (pH 11.0). Mam22 was effectively solubilized by 0.1 M Caps-NaOH buffer (pH 11.0) from the magnetosomes, whereas Mam12 remained in the magnetosomes. Precision Plus protein standards (Bio-Rad) were used (lane M).
Localizations of Mam22 and Mam12 in purified magnetosomes.
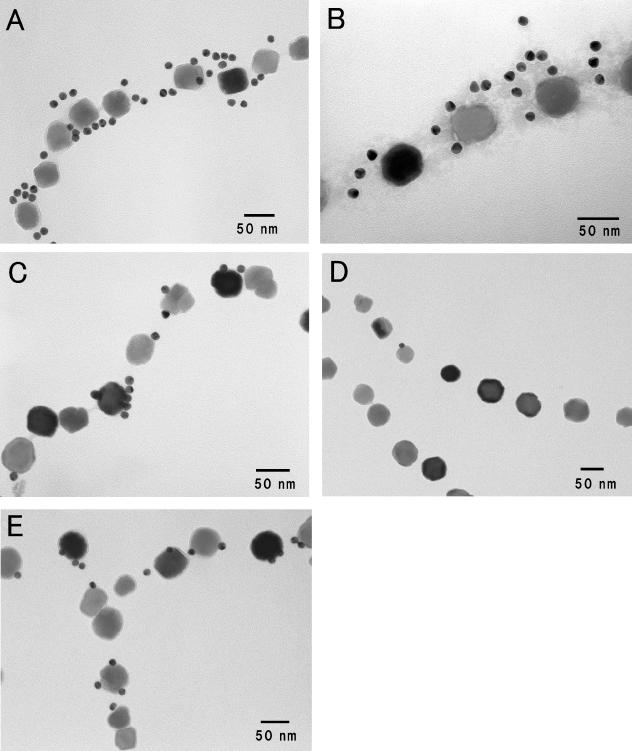
Immunogold staining was performed to visualize Mam22 and Mam12 in the magnetosomes using anti-Mam22 and anti-Mam12 polyclonal antibodies, which were generated as described in Materials and Methods. Each gold particle should represent a complex of Mam22 (Mam12), anti-Mam22 (Mam12), rabbit IgG, and anti-rabbit IgG antibody-gold conjugate. As shown in Fig. 3A and B, most Mam22 was localized on the magnetosomal matrix of the magnetosomes. Also, the result could not preclude the possibility that Mam22 was located on the interparticle connection, because it was difficult to discriminate spatially between the magnetosomal matrix and the interparticle connection, whereas the location of Mam12 was restricted to the surfaces of the magnetosomes (Fig. 3C). Very few gold particles were observed in the negative control experiment using preimmune serum (data not shown). On the other hand, the alkaline-treated magnetosomes remained in the original chain structures (Fig. 3D and E). However, as shown in Fig. 3D, most Mam22 was removed from the magnetosomes by the alkaline buffer treatment (Fig. 3D). It should be noted that Mam12 was not removed by the alkaline buffer treatment (Fig. 3E). These results are consistent with SDS-PAGE analyses of the control magnetosomes and the alkaline-treated magnetosomes (Fig. 2).
FIG. 3.
Immunogold labeling of Mam22 and Mam12 in purified magnetosomes. The magnetosomes were immunogold labeled using 15-nm gold particle-conjugated anti-Rabbit IgG for Mam22 or Mam12. (A) Localization of Mam22. (B) Localization of Mam22 with negative staining (2% sodium tungstate). (C) Localization of Mam12. (D) Localization of Mam22 after alkaline buffer treatment. (E) Localization of Mam12 after alkaline buffer treatment. Note that although most Mam22 was removed from the magnetosomes by the alkaline buffer treatment, Mam12 remained in the alkaline-treated magnetosomes.
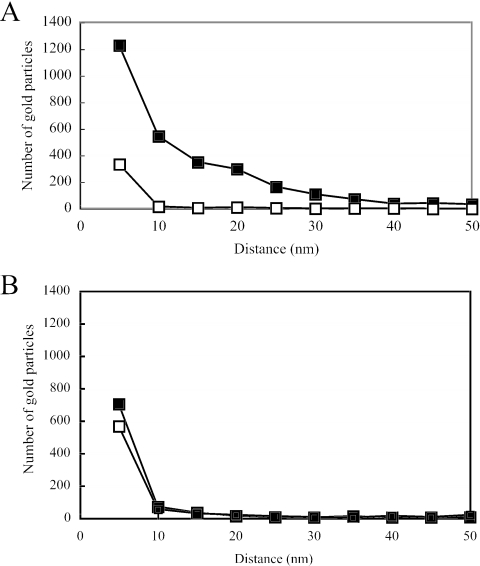
To more precisely clarify the distributions of Mam22 and Mam12 in the magnetosomes, the relationships between the numbers of gold particles and the distances from the surfaces of the gold particles to the surfaces of the magnetite particles were plotted. As shown in Fig. 4A, the distribution area of Mam22 in the intact magnetosomes reached 40 nm from the surfaces of the magnetite particles, while the number of gold particles was drastically decreased in the alkaline-treated magnetosomes. On the other hand, most Mam12 was distributed within 10 nm from the surfaces of the magnetite particles of the alkaline-treated magnetosomes, as in the control magnetosomes (Fig. 4B). Therefore, it is strongly indicated that most Mam22 is located in the magnetosomal matrix, while Mam12 is located in the magnetosomal membrane.
FIG. 4.
Relationship between the number of gold particles and the distance from the surfaces of gold particles to the surfaces of magnetite particles. Gold particles specific for Mam22 (A) and Mam12 (B) were used for immunogold staining with purified magnetosomes (solid squares) and alkaline-treated magnetosomes (open squares). The total number of gold particles in each plot was estimated from 1,000 magnetosome particles.
To definitely confirm the localization of Mam22, we examined the reconstitution of the alkaline-treated magnetosomes with the recombinant Mam22. As shown in Fig. 5A, the immunogold particles that bound to Mam22 were localized in the magnetosomal matrix. This result indicates that Mam22 reversibly binds to the magnetosomes in vitro.
FIG. 5.
Binding of the recombinant Mam22 to alkaline-treated magnetosomes. (A) Transmission electron micrograph of alkaline-treated magnetosomes incubated with recombinant Mam22. The 5-nm immunogold particles specific for Mam22 were observed around the magnetosomal matrix. (B) Transmission electron micrograph of alkaline-treated magnetosomes that were not incubated with recombinant Mam22 as a control. No gold particles were found around the magnetosomes.
Localizations of Mam22 and Mam12 in the cell.
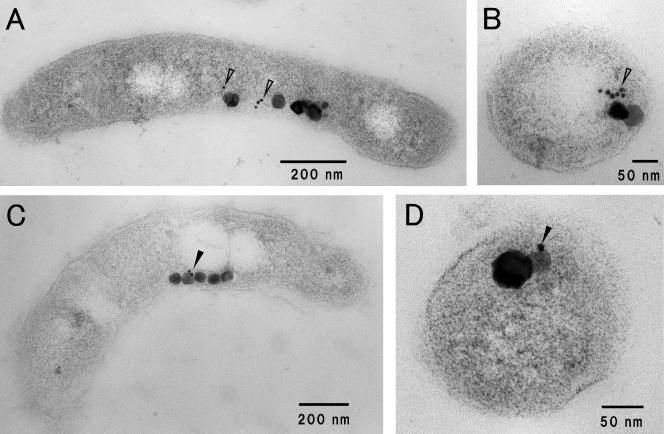
The localizations of Mam22 and Mam12 in M. magnetotacticum cells were examined by immunogold staining of ultrathin sections. The distributions of 5-nm gold particles on sections of the M. magnetotacticum cells showed that most Mam22 exists around and between the magnetosome particles (Fig. 6A and B), while Mam12 exists on the surfaces of magnetite particles (Fig. 6C and D). These results are consistent with those of the immunogold labeling of the purified magnetosomes, as shown in Fig. 3.
FIG. 6.
Localizations of Mam22 and Mam12 in M. magnetotacticum cells. Ultrathin sections prepared from M. magnetotacticum were labeled with 5-nm gold particles specific for Mam22 (A and B) and Mam12 (C and D). Note that Mam22 exists around and between magnetosome particles (open arrowheads), whereas Mam12 exists on the surfaces of magnetosome particles (solid arrowheads). Panels A and C are longitudinal transmission electron micrographs of the M. magnetotacticum cells. Panels B and D are transverse transmission electron micrographs of the M. magnetotacticum cells.
DISCUSSION
Previous studies reported the presence of a magnetosomal matrix that surrounds a chain of magnetite particles (6, 15, 23). An organic matrix of magnetosomes in M. magnetotacticum was removed by several rinses with 1.0 M NaCl solution (6). Taylor and Barry observed a semicrystalline matrix (magnetosomal matrix) with lattice fringes in uncultured magnetic bacteria (23). In the present study, we prepared the magnetosomes without NaCl and found a fine fibrous structure in the interparticle spacing of the magnetosome chain using the negative staining method; we also showed that a part of the structure around a line of magnetite particles was positively stained by uranyl acetate and sodium tungstate solution (Fig. 1). We propose that magnetosomes consist of four constituents, i.e., magnetite particle, magnetosomal membrane, magnetosomal matrix, and interparticle connection. Furthermore, we investigated the precise localizations of magnetosome-associated proteins, Mam22 and Mam12, on the fine structures of the magnetosomes by immunogold staining and showed that although Mam12 is localized on the magnetosomal membrane, Mam22 exists in the magnetosomal matrix.
It is known that magnetosomes of magnetic bacteria, such as M. magnetotacticum, M. gryphiswaldense, and Magnetospirillum sp. strain AMB-1, include various kinds of putative membrane proteins and soluble proteins, which are encoded by the mam gene island (7, 8, 14, 22). These molecular features of the magnetosome-associated proteins might be attributed to localization sites in the magnetosomes. In the present study, we used immunogold staining of the purified magnetosomes and ultrathin sections of the M. magnetotacticum cells and revealed that most Mam22 is localized on the magnetosomal matrix (Fig. 3 and Fig. 6). Furthermore, we prepared Mam22-deficient magnetosomes using alkaline solution, 0.1 M Caps-NaOH buffer (pH 11.0), and investigated the localization of Mam22 in vitro (Fig. 5). Interestingly, recombinant Mam22 is attached to the magnetosomal matrix of the alkaline-treated magnetosomes (Fig. 5). On the other hand, immunogold labeling against Mam12 showed that the protein is strictly localized on the magnetosomal membrane, but not on the magnetosomal matrix or interparticle connection. Therefore, it is suggested that the soluble magnetosome-associated proteins may be localized in the external constituents, such as the magnetosomal matrix and interparticle connection.
Recently, Komeili et al. generated the ΔmamA strain of Magnetospirillum sp. AMB-1 and proposed that MamA may play a part in magnetosome assembly and maintenance processes or regulation of the lengths of magnetosomes (10). However, the ΔmamA mutant is magnetic and has short-chain magnetosomes in the cell (10). Furthermore, Mam22-deficient magnetosomes retain the chain structure, as shown in Fig. 3D and E. Therefore, it is speculated that Mam22 does not participate in the maintenance of the chain structure of magnetosomes. Mam22 has five or six TPR motifs in the molecule (16, 17). It is generally accepted that the TPR motif mediates protein-protein interactions and the assembly of multiprotein complexes (5). Although Mam22 with TPR motifs might bind to the magnetosomal matrix using a protein-protein interaction, the Mam22 binding magnetosome-associated protein remains to be determined. Further studies are needed for the determination of a partner molecule of Mam22 in the magnetosome.
Very recently, Scheffel et al. observed a cytoskeleton-like filamentous structure extending up to the cell pole in M. gryphiswaldense cells by using cryoelectron tomography (21). Furthermore, the localizations of two Mam proteins, MamA (Mam22) (10) and MamJ (21), are also visualized as linear structures from pole to pole. Because Mam22 is a component of the magnetosomal matrix, this structure might be located from pole to pole of the cell and exceed the chain of magnetosomes. The magnetosomes have complex structures, which are constituted of various organic components; in particular, the magnetosome-associated proteins might have crucial functions for the biomineralization of magnetite and maintenance of the magnetosomes. The spatial localizations of the magnetosome-associated proteins in the magnetosome would provide useful information to elucidate the functional roles of these proteins.
Acknowledgments
This work was supported by a Human Frontier Science Program Research Grant (RGP0035/2004-C104) to Y.F. and a Grant-in-Aid for Scientific Research on Priority Areas (no. 16087205) to Y.F. from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
We thank T. Onoe (Taisei Gakuin University) for useful technical comments and K. Tazaki (Kanazawa University) for giving us the opportunity to use the TEM (JEM-2000EX). We also thank T. Fujiwara (Shizuoka University) for his helpful discussions.
REFERENCES
- 1.Arakaki, A., J. Webb, and T. Matsunaga. 2003. A novel protein tightly bound to bacterial magnetic particles in Magnetospirillum magneticum strain AMB-1. J. Biol. Chem. 278:8745-8750. [DOI] [PubMed] [Google Scholar]
- 2.Balkwill, D. L., D. Maratea, and R. P. Blakemore. 1980. Ultrastructure of a magnetotactic spirillum. J. Bacteriol. 141:1399-1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bazylinski, D. A., and R. B. Frankel. 2004. Magnetosome formation in prokaryotes. Nat. Rev. Microbiol. 2:217-230. [DOI] [PubMed] [Google Scholar]
- 4.Blakemore, R. P., D. Maratea, and R. S. Wolfe. 1979. Isolation and pure culture of a freshwater magnetic spirillum in chemically defined medium. J. Bacteriol. 140:720-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D'Andrea, L. D., and L. Regan. 2003. TPR proteins: the versatile helix. Trends Biochem. Sci. 28:655-662. [DOI] [PubMed] [Google Scholar]
- 6.Gorby, Y. A., T. J. Beveridge, and R. P. Blakemore. 1988. Characterization of the bacterial magnetosome membrane. J. Bacteriol. 170:834-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grünberg, K., E.-C. Müller, A. Otto, R. Reszka, D. Linder, M. Kube, R. Reinhardt, and D. Schüler. 2004. Biochemical and proteomic analysis of the magnetosome membrane in Magnetospirillum gryphiswaldense. Appl. Environ. Microbiol. 70:1040-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grünberg, K., C. Wawer, B. M. Tebo, and D. Schüler. 2001. A large gene cluster encoding several magnetosome proteins is conserved in different species of magnetotactic bacteria. Appl. Environ. Microbiol. 67:4573-4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnstone, A., and R. Thorpe. 1987. Immunochemistry in practice, 2nd ed. Blackwell Scientific Publications, Oxford, United Kingdom.
- 10.Komeili, A., H. Vali, T. J. Beveridge, and D. K. Newman. 2004. Magnetosome vesicles are present before magnetite formation, and MamA is required for their activation. Proc. Natl. Acad. Sci. USA 101:3839-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 12.Mann, S., J. P. Hannington, and R. J. P. Williams. 1986. Phospholipid vesicles as a model system for biomineralization. Nature 324:565-567. [DOI] [PubMed] [Google Scholar]
- 13.Matsunaga, T., and Y. Okamura. 2003. Genes and proteins involved in bacterial magnetic particle formation. Trends Microbiol. 11:536-541. [DOI] [PubMed] [Google Scholar]
- 14.Matsunaga, T., Y. Okamura, Y. Fukuda, A. T. Wahyudi, Y. Murase, and H. Takeyama. 2005. Complete genome sequence of the facultative anaerobic magnetotactic bacterium Magnetospirillum sp. strain AMB-1. DNA Res. 12:157-166. [DOI] [PubMed] [Google Scholar]
- 15.Meldrum, F. C., S. Mann, B. R. Heywood, R. B. Frankel, and D. A. Bazylinski. 1993. Electron microscopy study of magnetosomes in two cultured vibrioid magnetotactic bacteria. Proc. R. Soc. Lond. B. 251:237-242. [Google Scholar]
- 16.Okuda, Y., and Y. Fukumori. 2001. Expression and characterization of a magnetosome-associated protein, TPR-containing Mam22, in Escherichia coli. FEBS Lett. 491:169-173. [DOI] [PubMed] [Google Scholar]
- 17.Okuda, Y., K. Denda, and Y. Fukumori. 1996. Cloning and sequencing of a gene encoding a new member of the tetratricopeptide protein family from magnetosomes of Magnetospirillum magnetotacticum. Gene 171:99-102. [DOI] [PubMed] [Google Scholar]
- 18.Ouchterlony, O., and L. Ä. Nilsson. 1986. Immunodiffusion and immunoelectrophoresis, vol. 1, p. 32-50. In D. M. Weir, L. A. Herzenberg. C. Blackwell and L. A. Herzenberg (ed.), Handbook of experimental immunology, 4th ed. Blackwell Scientific Publications, Oxford, United Kingdom. [Google Scholar]
- 19.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 21.Scheffel, A., M. Gruska, D. Faivre, A. Linaroudis, J. M. Plitzko, and D. Schüler. 2005. An acidic protein aligns magnetosomes along a filamentous structure in magnetotactic bacteria. Nature 440:110-114. [DOI] [PubMed] [Google Scholar]
- 22.Schübbe, S., M. Kube, A. Scheffel, C. Wawer, U. Heyen, A. Meyerdierks, M. H. Madkour, F. Mayer, R. Reinhardt, and D. Schüler. 2003. Characterization of a spontaneous nonmagnetic mutant of Magnetospirillum gryphiswaldense reveals a large deletion comprising a putative magnetosome island. J. Bacteriol. 185:5779-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor, A. P., and J. C. Barry. 2004. Magnetosomal matrix: ultrafine structure may template biomineralization of magnetosomes. J. Microsc. 213:180-197. [DOI] [PubMed] [Google Scholar]
- 24.Ullrich, S., M. Kube, S. Schübbe, R. Reinhardt, and D. Schüler. 2005. A hypervariable 130-kilobase genomic region of Magnetospirillum gryphiswaldense comprises a magnetosome island which undergoes frequent rearrangements during stationary growth. J. Bacteriol. 187:7176-7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamazaki, T., H. Oyanagi, T. Fujiwara, and Y. Fukumori. 1995. Nitrite reductase from the magnetotactic bacterium Magnetospirillum magnetotacticum, a novel cytochrome cd1 with Fe(II):nitrite oxidoreductase activity. Eur. J. Biochem. 233:665-671. [DOI] [PubMed] [Google Scholar]