Abstract
“Roseburia inulinivorans” is an anaerobic polysaccharide-utilizing firmicute bacterium from the human colon that was identified as a producer of butyric acid during growth on glucose, starch, or inulin. R. inulinivorans A2-194 is also able to grow on the host-derived sugar fucose, following a lag period, producing propionate and propanol as additional fermentation products. A shotgun genomic microarray was constructed and used to investigate the switch in gene expression that is involved in changing from glucose to fucose utilization. This revealed a set of genes coding for fucose utilization, propanediol utilization, and the formation of propionate and propanol that are up-regulated during growth on fucose. These include homologues of genes that are implicated in polyhedral body formation in Salmonella enterica. Dehydration of the intermediate 1,2-propanediol involves an enzyme belonging to the new B12-independent glycerol dehydratase family, in contrast to S. enterica, which relies on a B12-dependent enzyme. A typical gram-positive agr-type quorum-sensing system was also up-regulated in R. inulinivorans during growth on fucose. Despite the lack of genome sequence information for this commensal bacterium, microarray analysis has provided a powerful tool for obtaining new information on its metabolic capabilities.
The human colon contains a dense and highly diverse microbial community consisting of over 500 different bacterial species (13, 15, 20, 41). These bacteria are predominantly obligate anaerobes and produce fermentation products that may be beneficial (e.g., butyrate) (35) or detrimental (e.g., hydrogen sulfide) (34) to the epithelial cells lining the human colon. The main butyrate-producing colonic anaerobes belong to Clostridium clusters IV and XIVa (3, 20, 26) and include cluster XIVa bacteria that have been assigned to the genus Roseburia (11). Studies using fluorescent in situ hybridization have shown that bacteria related to Roseburia (including Eubacterium rectale) can comprise up to 10% of the total bacterial population in human feces (21, 43). All Roseburia spp. produce butyrate, but they differ markedly in their substrate utilization profiles (12, 36).
Colonic bacteria gain energy from dietary substrates that escape digestion in the upper gastrointestinal tract, including plant cell wall polysaccharides, resistant starch, inulin, and a variety of oligosaccharides. In the distal colon, host-derived secretions and mucin, including the glycoproteins and glycolipids covering the surface of gut epithelial cells, may be significant substrates. l-fucose is frequently present at the terminus of epithelial glycoconjugates (14), and several pathogenic bacteria, including Salmonella enterica serovar Typhimurium LT2 (subsequently referred to as Salmonella serovar Typhimurium LT2) and the commensal bacterium Bacteroides thetaiotaomicron, utilize fucose for growth (see references 1 and 38, respectively). B. fragilis is able to incorporate fucose into its own surface polysaccharides, a mechanism that gives it a competitive advantage in colonizing a mouse intestine (8). Many other Bacteroides species, including B. thetaiotaomicron, are able to incorporate exogenous fucose directly into cellular glycoproteins (8).
Roseburia sp. strain A2-194 produces moderate amounts of butyrate in pure culture and was chosen for detailed study because of its ability to use a range of dietary substrates available in the colon for growth (12, 36). This strain is a representative of the predominant clostridial cluster XIVa group and belongs to the newly proposed species “Roseburia inulinivorans” (10). Unlike other Roseburia species, R. inulinivorans A2-194 is also able to grow on l-fucose, a sugar present in dietary polysaccharides such as pectin as well as in epithelial glycoconjugates. Here a genomic microarray from R. inulinivorans A2-194 was constructed and used to identify genes that are differentially expressed during growth on glucose and fucose. This shotgun microarray approach proved extremely powerful in revealing novel genes encoding degradative enzymes, substrate transport mechanisms, regulatory systems, and potential communication systems that respond to changes in growth substrates.
Studies of microbial gene expression that use microarray technologies have been largely confined to species for which complete genome sequences are available. Microarray technology can, however, be adapted to obtain valuable information on species whose genomes have not yet been sequenced. A shotgun microarray approach was first used to investigate the protozoal parasite Plasmodium falciparum (19) and has also been used to study gene expression in the gram-negative bacterium Leptospirillum ferrooxidans (33).
MATERIALS AND METHODS
Bacteria and growth conditions.
R. inulinivorans A2-194 (catalogue number NCIMB 14030 or DSM 16841) was routinely cultured in anaerobic M2GSC medium (29). Growth on single-carbon sources utilized anaerobic basal YCFA medium (12) supplemented with a 0.2% or 0.5% (wt/vol) concentration of the specific substrate (glucose, inulin [Dahlia], or fucose) as indicated. Strains were cultured in Bellco tubes under conditions of 100% CO2. Growth was determined spectrophotometrically by monitoring changes in optical density at 650 nm (OD650).
Fermentation product analyses.
Short-chain fatty acid production was determined by capillary gas chromatography (32) incorporating 2-ethyl butyric acid as an internal standard. Propanol production was also measured by gas chromatography using a Zebron ZBWax 30 m × 0.25 mm column to separate and quantify propanol and propanol, with butyraldehyde as an internal standard. All values represent the means of triplicate cultures.
RNA purification.
RNA was purified from midexponential phase cultures (OD650 = 0.5) on YCFA medium supplemented with a 0.5% concentration of either glucose or fucose by use of an RNeasy RNA purification kit (QIAGEN). Bacterial mRNA was further purified using a MICROBExpress kit (Ambion). This kit is designed to remove >95% of 16S and 23S rRNA from a total RNA purification, thereby effectively enriching the mRNA component. The manufacturers confirmed in silico that the kit would be effective in removing strain A2-194 16S rRNA from the total purified RNA volume. The purity and yield of total RNA and of MICROBExpress-treated RNA was confirmed using an Agilent 2100 bioanalyzer (Agilent Technologies).
DNA purification and cloning.
Total genomic DNA was purified from cultures grown on M2GSC by use of a genomic DNA extraction kit (Promega). DNA was partially restriction digested with Sau3AI, and the 1- to 2-kb-size fraction was purified from an agarose gel by electroelution and treatment with a Wizard cleanup kit (Promega). Purified DNA was ligated with BamHI-digested, alkaline phosphatase-treated pUC 13 (39). The resulting ligation mix was used to transform Escherichia coli XLI Blue cells. The transformation efficiency was 3 × 103 transformants/5 μl ligation, with a <15% background of nonrecombinant plasmids. To estimate the number of clones (N) to screen, the formula N = ln(1 − P)/ln(1 − 1/n), where n = the total genome size divided by the insert size (39).
Colony picking and insert amplification.
Positive-testing colonies were consolidated into 384-well microtiter plates by use of an automated colony picker (BioPick; BioRobotics) and cultured overnight in 70 μl LB containing μg ml−1 ampicillin. These broths were then split to form the master plate for storage at −80°C and the stocks for PCR amplification. Plasmid inserts were amplified by PCR using universal M13 primers, and the presence of single products was confirmed by agarose gel electrophoresis. A total of 7,000 clones were picked and amplified by PCR until 4,884 distinct clones containing single inserts 1 to 2 kb in size were identified. The PCR products from these clones were purified (Millipore Multiscreen PCR cleanup filter plates) and stored in 384-well microtiter plates.
Construction of the total genome microarray.
PCR products amplified from 4,884 clones were arrayed in triplicate on amino-silane-coated microscope slides (Corning GAPS) by use of a MicroGrid II TAS spotter (BioRobotics). Controls were included as follows: for positive controls, A2-194 genomic DNA and A2-194 16S rRNA amplified by PCR using the Universal eubacterial primers fD1 and rP2 (44), which give a 1.5-kb PCR product comparable in size to the amplicons of the genomic library; for negative controls, a 700-bp amplicon of the green fluorescent protein gene and several human gene-specific PCR products. These controls were randomly distributed on the array. Landmark spots were positioned at strategic points on the microarray slide to facilitate orientation following hybridization (SpotReport Array validation system; Stratagene). Following microarray printing, the slides were baked at 80°C for 2 h and stored at room temperature.
RNA labeling.
MICROBExpress-treated RNA (1 μg) was labeled during cDNA synthesis by random nonamer extension incorporating either dCTP-Cy3 or dCTP-Cy5 dyes in the reverse-transcription (RT) step (CyScribe first-strand cDNA labeling kit; Amersham), and labeled products were cleaned up using a CyScribe GFX purification kit (Amersham). The RNA purification was spiked with SpotReport mRNA spikes 1, 2, and 3 (Stratagene) prior to labeling to enable orientation of the array following hybridization.
Microarray hybridization.
Hybridization was done using a GeneTAC hybridization station (Genomic Solutions) and agitation and circulating buffers. Slides were first blocked in buffer (180 mM succinic anhydride, 44 mM sodium borate in 1-methyl-2-pyrrolidone) and then washed in deionized water at 98°C (twice for 1 min each time) and finally in 95% ethanol (1 min) at room temperature. After drying, the slides were prehybridized at 50°C for 30 min (in 10 mg ml−1 fraction V bovine serum albumin, 3.5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], and 0.1% sodium dodecyl sulfate [SDS]), washed again in deionized water, dried, and immediately hybridized. The hybridization solution [labeled cDNA mixed with 10 μg human Cot-1 DNA (Invitrogen), 15.4 μg Saccharomyces cerevisiae tRNA, 8 μl poly(dA), 14.4 μl 20× SSC, and 2.4 μl 100× Denhardt's solution] was first boiled for 2 min and cooled to 45°C before addition of 1.4 μl 5% SDS and incubation at 65°C for 30 min. This solution was then added to the array slides, which were hybridized for 16 h at 65°C before being washed at room temperature in medium-stringency buffer (0.1% SDS, 0.5× SSC) for 5 min followed by two high-stringency washes (0.1% SDS, 0.1× SSC) of 5 min each.
The dye labeling was swapped for a second hybridization, and a separate RNA purification volume was also labeled and hybridized twice to ensure reproducibility and to obtain statistically significant results.
Scanning and data analysis.
In total, four slides were hybridized for each comparison, making a total of 12 hybridizing spots per amplified clone. The fluorescence of each spot was measured in two channels using a GeneTAC LS IV hybridization station (Genomic Solutions) and GeneTac Integrator version 3.0.1 software. Spot intensities were log transformed, and Loess normalization (46) was applied to remove differences in probe labeling and hybridization efficiencies. One-sample t tests were applied to the log-ratio values to test for differential expression. Due to the small number of replicates (two independent samples) and the lack of parametric assumptions, the corresponding P values were regarded as a rough indicator of importance. In this study we thus ranked clones based on their average fold change results and selected those that had a P value of <0.2 and showed at least a fourfold increase in induction consistently across all hybridizing spots.
DNA sequence analysis.
Plasmids were purified from positive-testing clones (Promega Wizard miniprep kit) and the inserts sequenced using a CEQ 8000 genetic analysis system (Beckman Coulter). Sequences were screened for database homologues using the BLAST search tool (http://www.ncbi.nlm.nih.gov/BLAST/). Sequences of interesting genes were extended using internal primers to complete the sequence of the entire insert, and genome walking (Clontech Universal Genome Walker kit) on purified genomic DNA was used to extend the sequences up and/or downstream of the cloned DNA. Sequences were assembled initially using the UWGCG program (9) available through the HGMP facility or latterly the Lasergene version 6 program (DNAStar, Inc).
Real-time RT-PCR.
Primers were designed to specifically amplify fragments of 80 to 200 bp from selected clones. The optimal annealing temperature for each primer pair was confirmed using a standard PCR amplification in a gradient PCR machine (Bio-Rad) capable of running 10 different annealing temperatures simultaneously using the appropriate clone DNA as a template. The RNA purification mixture (10 ng) used to hybridize to the array slides was reverse transcribed using an iScript cDNA synthesis kit (Bio-Rad). The resulting cDNA (2 μl) was used in real-time PCR amplifications, utilizing the Bio-Rad iCycler iQ and incorporating iQ SYBR green Supermix (Bio-Rad) according to the manufacturer's recommendations. Control samples lacking the reverse transcriptase enzyme were included to assess DNA contamination of the RNA sample, and triplicate samples of a fivefold dilution series were amplified in each case. Amplification conditions were as follows: initial denaturation at 95°C for 3 min; 40 cycles of 95°C for 30 s and the specific annealing temperature for 30 s; and a melting cycle from 52°C to 95°C. Threshold cycle (Ct) values for fucose and glucose RNA, obtained in the same run, were compared to determine the gene induction level, after conversion from a logarithmic to a linear scale using the formula x = 2−Ct. In all cases plasmid controls were included to form standard curves, and the final melt curve analysis step confirmed the specificity of amplification.
Nucleotide sequence accession numbers.
These sequence data have been submitted to the GenBank database under database accession numbers DQ299881 (p4p5), DQ299882 (contig 6), DQ299883 (contig 3), and DQ299884 (contig 1-5-2).
RESULTS
Adaptation of R. inulinivorans to growth on fucose.
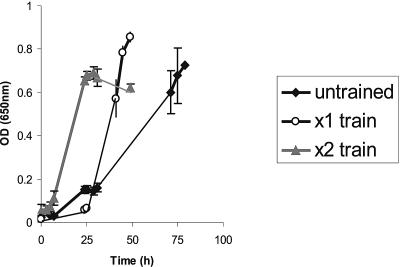
R. inulinivorans A2-194 was able to grow on basal YCFA medium supplemented with 0.5% l-fucose as a sole carbon source for growth. Cultures transferred from glucose-containing medium grew slowly on fucose after a lengthy (48-h) lag phase, reaching an OD650 of 0.60 after 72 h. However, two periods of successive subculturing in fucose-containing medium decreased the lag period and cultures achieved the same OD after 24 h (Fig. 1). Cells thus “adapted” to growth on fucose were confirmed to be R. inulinivorans A2-194 by 16S rRNA sequencing and are referred to as R. inulinivorans A2-194(F). The lag is assumed to result from the need to induce functions required for the fucose utilization; there was no evidence for mutational adaptation, since, following revival from −80°C storage, cultures previously able to grow well on fucose showed the same pattern of delayed growth and decreasing lag periods with successive subcultivations on fucose. Cultures that were exposed to growth on fucose for extended periods formed the most effective inoculants, with subsequent cultures growing most rapidly to the highest ODs (data not shown).
FIG. 1.
Growth of R. inulinivorans A2-194 on fucose monitored by measuring OD650. Untrained culture 1 was used as the inoculum for culture 2 (×1 train), which was the inoculum for culture 3 (×2 train). Inoculations were done after 24 h growth of the preceding culture. Results represent the averages of triplicate growth tube results; standard deviation bars are shown.
During growth on glucose or inulin, R. inulinivorans A2-194 produces butyrate (∼8 mM) as its main fermentation product and produces little propionate. When the strain was grown on 0.5% fucose, however, propionate, propanol, and butyrate were detected in culture supernatant of the fucose-adapted A2-194(F) culture after 16 h (Table 1). There was no difference in the fermentation products released by R. inulinivorans A2-194 or A2-194(F) on glucose or inulin substrates, and no propanol was detected. Neither culture was able to grow on 1,2-propanediol alone, but propanol and propionate were produced from 1,2-propanediol in the presence of glucose (Table 1).
TABLE 1.
Fermentation products of R. inulinivorans on different growth substrates
| Substrate and straina | Concn of fermentation product (mM ± SD)b
|
||
|---|---|---|---|
| Propionate | Propanolc | Butyrate | |
| Glucose (0.2%) | |||
| A2-194 | 1.19 ± 0.12 | 0 | 8.34 ± 0.12 |
| A2-194(F) | 0.15 ± 0.27 | 0 | 8.20 ± 0.64 |
| Inulin (0.5%) | |||
| A2-194 | 0.17 ± 0.12 | 0 | 8.25 ± 1.08 |
| A2-194(F) | 0.5 ± 0.16 | 0 | 8.88 ± 1.90 |
| Glucose (0.2%) + 1,2- propanediold | |||
| A2-194 | 1.29 ± 0.08 | 9.29 ± 1.32 | 7.10 ± 0.02 |
| A2-194(F) | 3.88 ± 0.36 | 22.16 ± 6.45 | 14.11 ± 0.04 |
| Fucose (0.5%) | |||
| A2-194e | |||
| A2-194(F) | 18.84 ± 0.64 | 7.31 ± 1.72 | 12.77 ± 0.93 |
R. inulinivorans A2-194, wild-type strain; A2-194(F), strain able to grow on fucose.
Values shown are means ± standard deviations of the results for triplicate growth tubes cultured for 16 h on YCFA medium with specific supplements.
Propanol production was negligible on glucose and inulin.
Growth on 1,2-propanediol (82 mM) was only possible with glucose supplementation.
Strain A2-194 grows on fucose only after an extended lag phase (illustrated in Fig. 1).
Microarray hybridization and identification of differentially expressed clones.
Pulsed-field gel electrophoresis analysis had indicated that the size of the genome of R. inulinivorans A2-194 was at least 2 Mb (data not shown). Consequently, screening 4,673 random clones with 1- to 2-kb inserts would give an estimated genome coverage of 97% (39). Recombinant clones containing single inserts of appropriate size were identified, and the PCR products from 4,884 of these clones were arrayed in triplicate as described in Materials and Methods. The total number of spots on a single microarray slide, including controls and landmarks, was approximately 15,000. The replication and controls included on the array enabled statistically reliable data to be produced.
Bacterial mRNA was purified on two separate occasions from midexponential-phase cultures grown on either glucose or fucose. Aliquots of pairs of mRNA samples were hybridized to different microarray slides, incorporating a dye swap (e.g., for slide 1, glucose Cy3 and fucose Cy5, and for slide 2, glucose Cy5 and fucose Cy3) to account for any differences in dye incorporation. Relatively few transcripts were expressed on both fucose and glucose, indicative of a major switch in bacterial metabolism.
Following statistical analysis, clones were ranked based on the degree of induction, and the order was confirmed by determining P values. A total of 80 clones showed at least fourfold increases in induction on fucose compared to glucose and the increases of the five most strongly induced clones were between 30- and 60-fold. Inserts were sequenced for the top 25 clones (corresponding to >eightfold induction) and for 10 additional clones induced between four- and eightfold, all with P values of <0.2. The best sequence matches based on BLASTX searches are shown elsewhere (see Table 1S in the supplemental material). In contrast, only 5 clones showed >10-fold repression on fucose compared to glucose, and the 10 most repressed clones were also sequenced (Table 1S).
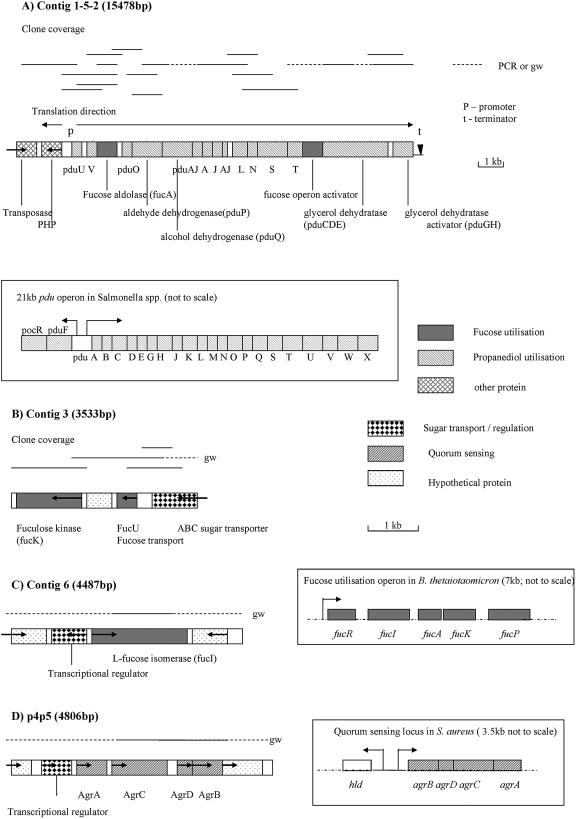
Many of the sequences for highly induced clones overlapped, forming six contigs between 3 and 6 kb in length. Extension of some sequences by genome walking demonstrated that three of these contigs fell into a single large region of DNA > 15 kb in size (contig 1-5-2). The four longest contigs analyzed are illustrated diagrammatically in Fig. 2. Full-length open reading frames (ORFs) carried on these contigs were searched against the database (BLASTP search) to assign functions to the proteins. Between them, these contigs contain candidate genes carrying out most of the steps reported to be involved in the anaerobic metabolism of fucose to propionate and propanol in other bacteria (Fig. 2, Fig. 3, Table 2).
FIG. 2.
Arrangements of ORFs involved in fucose utilization in the four major contigs assembled following microarray screening. The direction of translation is indicated. The positions of the hybridizing clones are given above the main diagram and the regions extended by genome walking (gw) or PCR amplification are indicated by dashed lines. The arrangement of related, published operons of genes is indicated within boxes. The R. inulinivorans A2-194 contigs are drawn to scale (scale bars shown). Note that the size of the scale for contig 1-5-2 is half that of the other contigs. (A) Contig 1-5-2, propanediol utilization genes, and the corresponding Salmonella serovar Typhimurium LT2 pdu operon (4); (B) contig 3, fuculose kinase, and adjacent genes; (C) contig 6, fucose isomerase and surrounding genes, and the fucose utilization operon from B. thetaiotaomicron (23); (D) p4p5, quorum-sensing operon and the corresponding S. aureus quorum-sensing operon (30).
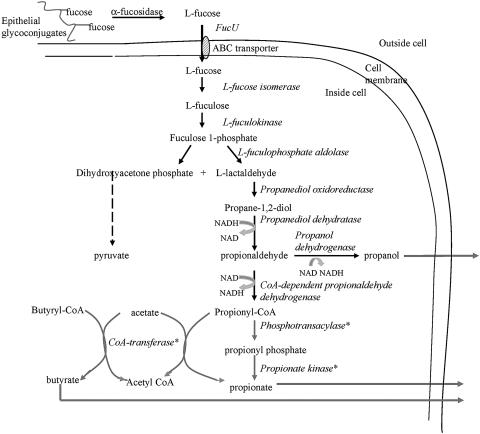
FIG. 3.
Schematic illustrating the predicted pathway for anaerobic fucose metabolism by R. inulinivorans A2-194. Enzyme names are shown in italics. The possible alternative conversion routes to form propionate are shown with asterisks.
TABLE 2.
Summary of the different ORFs present in the major contigs assembled following the fucose screen
| Contig or clone | Strain A2-194 protein
|
Related protein
|
||||||
|---|---|---|---|---|---|---|---|---|
| ORF | ORF length (in amino acids) | Predicted function of protein in strain A2-194 | Functional equivalenta | % Identity | Name of closest relative | Closest species | E value | |
| Contig 1-5-2 | 1 | 244b | Putative transposase | 35 | Putative transposase | Clostridium thermocellum | 5e-36 | |
| 2 | 282 | Histidinol phosphatase | 35 | PHP family proteinc | Clostridium acetobutylicum | 2e-33 | ||
| 3 | 117 | Polyhedral bodies | PduU | 55 | Propanediol utilization protein PduU | Clostridium perfringens | 3e-30 | |
| 4 | 147 | Unknown | PduV | 42 | Propanediol utilization protein PduV | Salmonella enterica | 2e-23 | |
| 5 | 264 | l-Fuculose aldolase | FucA | 35 | l-Fuculose phosphate aldolase | Rhodopirellula baltica | 3e-45 | |
| 6 | 118 | Polyhedral bodies | PduO? | 48 | Uncharacterized Pdu protein (36% PduO) | Rhodospirillum rubrum | 2e-23 | |
| 7 | 457 | Propionaldehyde dehydrogenase | PduP | 48 | CoA-dependent propionaldehyde dehydrogenase | Listeria monocytogenes | 1e-108 | |
| 8 | 403 | Putative propanol dehydrogenase | PduQ | 48 | Zn-dependent alcohol dehydrogenase | Oenococcus oeni | 6e-96 | |
| 9 | 112 | Polyhedral bodies | PduA | 35 | Polyhedral body shell protein PduA | Listeria monocytogenes | 2e-06 | |
| 10 | 93 | Polyhedral bodies | PduJ | 80 | Polyhedral body shell protein PduJ | Salmonella enterica | 2e-19 | |
| 11 | 118 | Polyhedral bodies | PduJ | 38 | Carboxysome protein | Desulfitobacterium hafniense | 5e-10 | |
| 12 | 94 | Polyhedral bodies | PduA | 72 | Polyhedral body shell protein PduA | Listeria monocytogenes | 9e-20 | |
| 13 | 203 | Unknown | PduL | 50 | Propanediol utilization protein PduL | Desulfitobacterium hafniense | 4e-46 | |
| 14 | 90 | Polyhedral bodies | PduN | 51 | Carboxysome protein | Desulfitobacterium hafniense | 1e-15 | |
| 15 | 441 | NADH oxidoreductase | PduS? | 41 | Predicted oxidoreductase (38% pduS) | Desulfitobacterium hafniense | 5e-89 | |
| 16 | 181 | Polyhedral bodies | PduT | 39 | Carboxysome protein | Desulfitobacterium hafniense | 6e-32 | |
| 17 | 259 | Transcriptional regulator | 41 | Transcriptional regulator (of sugar metabolism) | Oceanobacillus iheyensis | 3e-45 | ||
| 18 | 841 | Propanediol dehydratase | PduCDE | 53 | B12-independent glycerol dehydratase | Clostridium butyricum | 0.0 | |
| 19 | 264 | Propanediol dehydratase activator | PduGH | 35 | Glycerol dehydratase activator | Clostridium butyricum | 3e-53 | |
| Contig 3 | 1 | 464 | l-Fuculose kinase | FucK | 40 | Rhamnulokinase/ l-fuculose kinase | Bacteroides thetaiotaomicron | 7e-92 |
| 2 | 144 | Hypothetical protein | No match | |||||
| 3 | 147 | Fucose transport | FucU | 49 | Fucose operon protein, FucU | Streptococcus pneumoniae | 8e-34 | |
| 4 | 287b | ABC sugar transporter | 31 | ABC sugar transporter | various | 2e-23 | ||
| Clone p4p5 | 1 | 270 | Putative outer membrane protein | 23 | Secreted protein with vWA domaind | Streptomyces coelicolor | 0.003 | |
| 2 | 199 | AgrB | AgrB | 29 | Accessory gene regulator AgrB | Staphylococcus aureus | 0.072 | |
| 3 | 50 | Predicted AIP peptide precursor | AgrD | No match | ||||
| 4 | 431 | Histidine kinase ATPase | AgrC | 32 | Histidine kinase related ATPase | Treponema denticola | 1e-21 | |
| 5 | 219 | Two-component response regulator | AgrA | 28 | Two-component response regulator | Enterococcus faecalis | 3e-24 | |
| 6 | 113 | Transcriptional regulator | 35 | Predicted transcriptional regulator | Clostridium acetobutylicum | 1e-11 | ||
| 7 | 124 | Hypothetical protein | No match | |||||
| Contig 6 | 1 | 236b | Zn-dependent alcohol dehydrogenase | 24 | Alcohol dehydrogenase (zinc) | Pyrobaculum aerophilum | 0.44 | |
| 2 | 257 | Transcriptional regulator | 24 | AraC/XylS transcriptional regulator | Bacillus halodurans | 3e-15 | ||
| 3 | 599 | l-Fucose isomerase | FucI | 61 | l-Fucose isomerase | Bacteroides thetaiotaomicron | 0.0 | |
| 4 | 170 | Hypothetical protein | No match | |||||
Pdu, propanediol utilization nomenclature based on that of Salmonella serovar Typhimurium LT2; Fuc, fucose utilization nomenclature based on that of E. coli; Agr, quorum-sensing system from S. aureus.
Incomplete ORF.
PHP, phosphohistidinol phosphatase.
vWA, von Willebrand factor type A domain.
Three of the most repressed clones corresponded to clones that had been shown to be inducible on inulin in a separate study (data not shown). Two of them overlapped and encoded a galactoside transport ATP binding protein. Another three clones encoded enzymes involved in the transfer of the acetyl-coenzyme A (CoA) group between formate and pyruvate, a key step in anaerobic glucose metabolism (see Table 1S in the supplemental material).
Genes induced during growth on fucose relative to glucose.
Three fucose-inducible genes were detected whose products are predicted to catalyze the intracellular conversion of l-fucose to l-lactaldehyde. In fact, the fucose isomerase (contig 6) and fuculose kinase (contig 3) genes were the most highly induced genes detected (58- and 54-fold, respectively), with the fuculose aldolase ranked fifth (33-fold). The fuculose isomerase and fuculose kinase genes had greatest identity to homologues from B. thetaiotaomicron, whereas the aldolase gene was most similar to a gene from a different bacterium (Table 2). These genes were not adjacent to each other in R. inulinivorans, although in other bacteria (e.g., B. thetaiotaomicron, E. coli, and Salmonella spp.) they occur together in a single operon that includes a repressor gene. In B. thetaiotaomicron a fucose permease transport protein is encoded slightly downstream of the three degradative genes and has its own promoter (23). A fucose permease gene was not identified in R. inulinivorans, but two genes upstream of the fuculose kinase in contig 3 encoded homologues of sugar transporters (Fig. 2, Table 2). One of these had 49% identity to the FucU fucose transport protein from Salmonella pneumoniae, and the other 31% identity to various sugar transporters.
All but two of the R. inulinivorans A2-194(F) clones whose sequences fell into contig 1-5-2 were induced more than 10-fold during growth on fucose. This contig contained 19 ORFs, 15 of which were fucose-inducible homologues of propanediol utilization (pdu) genes, one of which was the fuculose aldolase homologue (Fig. 2, Table 2), and one of which had identity to a fucose operon activator. The first two ORFs in this contig encode proteins with no obvious function in fucose degradation. Both these sequences were contained within a single clone, and it is likely that the sequence at the other end of the clone (pduU) was responsible for the hybridization signal. Separating these two genes from the remaining ORFs in contig 1-5-2 are 367 nucleotides (nt) of noncoding sequence that contain two putative promoter regions presumed to be responsible for the divergent transcription of the histidinol phosphatase and the pduU and subsequent genes (Fig. 2). A 20-nt inverted repeat sequence forming a strong putative terminator sequence (ΔG = −13.3 kcal; melting temperature = 78.7°C) is present 40 nt downstream of the final ORF in this contig encoding the putative propanediol dehydratase activator.
In Salmonella serovar Typhimurium LT2, 23 pdu genes are organized into a gene cluster. These include genes that encode activities involved in the conversion of 1,2-propanediol to propionate or propanol and also genes that encode proteins required for polyhedral body formation (18). Polyhedral bodies are analogous to carboxysomes and are assumed to keep the toxic propionaldehyde intermediate formed by the conversion of 1,2-propanediol to propionyl CoA sequestered away from the rest of the cell (18). The order of the gene homologues in R. inulinivorans A2-194 is, however, quite different from that found in Salmonella serovar Typhimurium LT2.
Of the four major structural polyhedral body proteins (PduJ, PduA, PduB′, PduB) in Salmonella serovar Typhimurium LT2, only analogues of PduA and PduJ (themselves 82% identical) (4) are present in R. inulinivorans A2-194, although there are four contiguous genes whose closest relative is either pduA or pduJ (Table 2). Two of these genes (carried by ORFs 10 and 12) are 77% identical to each other. Homologues of pduP and pduQ, encoding propionaldehyde dehydrogenase and propanol dehydrogenase enzymes, respectively, were detected in R. inulinivorans. No close homologues of pduCDEGH genes encoding a B12-dependent propanediol dehydratase enzyme were detected, although genes related to the C. butyricum B12-independent glycerol dehydratase gene, and its associated activator, were present. Since R. inulinivorans was unable to grow on glycerol and since these genes are linked to pdu homologues, they are assumed to be responsible for propanediol dehydratase activity.
Also induced (3.44-fold) on fucose was a transcript with sequence identity to the Staphylococcus aureus quorum-sensing Agr (accessory gene regulator) locus involved in density-dependent cell-to-cell signaling (Fig. 2; Table 2). The percentages of identity for each of the individual proteins in this operon were quite low (28% to 35%; Table 2), but in general the sequences of quorum-sensing regions are highly variable and highly specific for the host bacterium (28). The genes identified exist as a cluster of four, each with considerable sequence, size, and predicted secondary structure similarity to those of S. aureus (47). The R. inulinivorans AgrB and AgrC proteins both have five to six predicted membrane-spanning hydrophobic domains capable of interacting with the cell membrane and very similar in length and position to those in the corresponding S. aureus proteins (see references 48 and 5, respectively) (data not shown). The AgrD protein, the putative precursor of the active autoinducer octapeptide (AIP) molecule, has an N-terminal α-helical amphipathic motif. In S. aureus this motif is postulated to anchor the AgrD to the cell membrane, stabilizing it for processing and release of the mature AIP (49). Induction of agrB and agrC during growth on fucose was confirmed by RT-PCR (Table 3).
TABLE 3.
Comparison of gene expression measurements determined by microarray and RT-PCR
| Clone | Gene or protein | Fold changea
|
|
|---|---|---|---|
| RT-PCR | Microarray | ||
| p1k11 | Fucose isomerase | 123 | 58.4 |
| p1g6 | Fucose aldolase | 109 | 32.6 |
| p1g6 | pduV | 16.2 | 32.6 |
| p4e23 | fucU | 8.2 | 21.3 |
| p4p5 | agrB | 5.8 | 3.4 |
| p4p5 | agrC | 2.5 | 3.4 |
| p10n3 | Formate acetyl transferase | −11.9 | −12.1 |
| p7k5 | Galactoside transport protein | −3.2 | −19.5 |
The severalfold changes for gene expression on fucose relative to glucose. Both RT-PCR and microarray values have been log transformed (in base 2).
Analysis of specific changes by real-time PCR.
The differential regulation of selected ORFs was confirmed by quantitative real-time RT-PCR using RNA templates isolated from cells grown on either glucose or fucose medium. ORFs were chosen representing highly and moderately expressed and repressed genes. The severalfold induction or repression of each gene relative to the glucose standard was calculated and compared to the microarray data (Table 3). All the genes detected as induced on the microarrays were found by the RT-PCR analysis to be up-regulated, while those genes repressed on the microarray also showed negative fold changes in expression following RT-PCR (Table 3).
Constitutive α-fucosidase expression.
An exogenous α-fucosidase is required to release terminal fucose residues from pectin and epithelial glycoconjugates prior to uptake by the bacterial cell, but corresponding sequences were not detected by the microarray analysis. R. inulinivorans A2-194(F) cells grown on either fucose or glucose showed α-fucosidase activity, visualized by clear zone formation on plates containing 4-methylumbelliferyl-α-l-fucopyranoside (MUF; data not shown). This indicates that α-fucosidase activity is expressed constitutively, as it is in B. thetaiotaomicron (23).
DISCUSSION
The shotgun microarray analysis described here offers a powerful method for investigating gene expression that is applicable to any cultured bacterium. This is the first report of the use of this approach to investigate gene expression in a predominant commensal human gut bacterium. The present work identified transcriptional regulators, transport systems, and enzymes involved in fucose utilization in R. inulinivorans A2-194. R. inulinivorans A2-194, in common with several pathogenic bacteria and also B. thetaiotaomicron, is able to grow on fucose because it possesses inducible fucose utilization genes. The observed induction of these genes, and the resultant dramatic switch in metabolism, presumably explains the lag phase that R. inulinivorans A2-194 undergoes when transferred to growth medium containing fucose as the sole energy source. Metabolism of fucose produces propionate, propanol, and butyrate, while butyrate is the main product of R. inulinivorans A2-194 grown on glucose.
Anaerobic fucose metabolism integrates three metabolic pathways (Fig. 3), and the expression of genes corresponding to most of the enzymes involved in the conversion of l-fucose to propionate and propanol was induced on fucose. The enzymes involved in the initial uptake and conversion of fucose to l-lactaldehyde in R. inulinivorans A2-194 correspond to those identified in B. thetaiotaomicron and E. coli, where the relevant genes are clustered into operons. In common with B. thetaiotaomicron (23), the exogenous α-fucosidase is constitutively expressed and is required to release the fucose residue from the epithelial glycoconjugates for import into the bacterial cell. The l-lactaldehyde is reduced to 1,2-propanediol, which is excreted and lost from B. thetaiotaomicron and E. coli cells (16). In contrast, R. inulinivorans A2-194 is able to ferment the 1,2-propanediol into propionate and propanol, providing a sink for reducing equivalents and providing energy for the cell. Both of these end products were detected during growth on fucose. The conversion of l-lactaldehyde to 1,2-propanediol is normally carried out by propanediol oxidoreductase. Although a specific propanediol oxidoreductase homologue was not identified here, this step requires an NADH cofactor and thus may be carried out by one of the other enzymes, for example, the NADH oxidoreductase (ORF 15 on contig 1-5-2).
Propanediol utilization genes have been identified in pathogenic enteric bacteria (e.g., Salmonella spp., C. perfringens, C. tetani, and some Listeria spp.) but have not previously been associated with commensal gastrointestinal bacteria. The 20-kb propanediol utilization (pdu) operon has been fully characterized in Salmonella serovar Typhimurium LT2, where expression of 23 distinct genes is tightly regulated by an upstream AraC family transcription regulator (7). In R. inulinivorans A2-194 the genes involved in the fucose and pdu pathways appear to be interspersed, illustrating the interdependence of the metabolic routes. Most of the pdu genes do, however, appear to be carried on an operon with identifiable promoter and terminator sequences flanking the collection of ORFs. An ORF with significant identity to a transcriptional regulator (ORF 17, contig 1-5-2) is present within the gene cluster (Table 2). The method by which expression of these genes is controlled has not been elucidated and might involve glucose repression as well as induction by fucose.
No enzyme carrying out the final conversion of propionyl CoA to propionyl phosphate was identified, although an enzyme with identity to a hydroxyacetone kinase may act as the propionate kinase, producing propionate. Bobik et al. (4) did not identify a phosphotransacylase homologue in Salmonella serovar Typhimurium LT2. It may be, however, that this conversion occurs by a different route in R. inulinivorans and that it involves a CoA transferase and either acetate or butyrate as an alternative substrate (Fig. 3). Although a gene encoding a propionyl CoA transferase could not be detected in R. inulinivorans by use of degenerate PCR primers (P. Louis, personal communication), the bacterium does possess a butyryl CoA transferase (27), and the same enzyme in a related Roseburia sp. can use either propionate or butyrate as a cosubstrate (6).
Due to the toxic nature of the propionaldehyde intermediate, the conversion of 1,2-propanediol to propionyl CoA in Salmonella serovar Typhimurium LT2 is carried out within polyhedral bodies. Fifteen different proteins, of which 7 are structural, are associated with these polyhedral bodies (18). ORFs 9 to 12 of the R. inulinivorans pdu operon encode proteins closely matching PduA and -J, the two most abundant polyhedral body structural proteins from Salmonella serovar Typhimurium LT2 (18). The increased production of propanol relative to propionate in cells grown in the presence of 1,2-propanediol may be a mechanism to remove the toxic intermediate propionaldehyde from polyhedral bodies within the cell as quickly as possible. Propionaldehyde was barely detectable in cells during growth on either 1,2-propanediol plus glucose or on fucose.
Interesting evolutionary questions arise concerning the nature and distribution of genes for fucose utilization in gut bacteria. At least three types of fucose-utilizing systems apparently exist. Most strains of E. coli, and also of Bacteroides thetaiotaomicron 5482 (45), lack pdu genes required to utilize 1,2-propanediol (24). In these bacteria any 1,2-propanediol produced from fucose is assumed to be excreted from the cell, although its immediate precursor l-lactaldehyde can be converted under aerobic conditions to pyruvate via l-lactate. This allows energy to be derived from all of the fucose carbon skeleton under aerobic conditions, whereas half of it is lost under anaerobic conditions (2, 25). It is proposed that during evolution most enterobacteria lost the ability to metabolize 1,2-propanediol anaerobically. Salmonella serovar Typhimurium LT2, however, has regained this ability as a result of horizontal gene transfer (25). Salmonella serovar Typhimurium LT2 possesses a full complement of propanediol utilization genes that include a vitamin B12-dependent propanediol dehydratase plus the full complement of vitamin B12 synthetic genes required to supply this cofactor, which are contiguous on the genome and coregulated in response to the presence of fucose.
The third system is that for R. inulinivorans as reported here. The interspersed nature of the genes makes it unlikely that the pdu operon was acquired by R. inulinivorans A2-194 via a single horizontal gene transfer event. These proteins are predicted to catalyze the same pathway of reactions for propanediol utilization as found in Salmonella spp. but with the critical difference that the 1,2-propanediol dehydratase is a vitamin B12-independent type, with closest identity to a coenzyme B12-independent glycerol dehydratase and its activator enzyme from C. butyricum (37). The C. butyricum glycerol dehydratase enzyme also has activity against propanediol (31) and is thus far the only such enzyme fully described in the literature. An oxygen-sensitive diol dehydratase enzyme from C. glycolicum, described previously, was inducible and active against 1,2-propanediol and 1,2-ethanediol (17), but the sequence of this enzyme is not available. The reason why Salmonella should rely instead on a B12-dependent system is not known. Since the corresponding B12-independent glycerol dehydratase in C. butyricum is a highly oxygen-sensitive enzyme it might be that the B12-independent system is limited to strict anaerobes. The vitamin B12-independent propanediol dehydratase reported here could represent the typical mechanism for fucose utilization in Clostridium-related strict anaerobes, which make up the largest group of human colonic bacteria (26). Future work will focus on determining whether the R. inulinivorans enzyme is specific for 1,2-propanediol or whether it also has activity against other substrates, including glycerol and 1,2-ethanediol.
The identification of a possible quorum-sensing system in R. inulinivorans responding to the presence of fucose is also of particular interest. Colonization of mucins, gut epithelial surfaces, or pectin (all fucose-containing substrates) is likely to involve biofilm formation at a critical cell density, thus requiring a bacterial quorum-sensing mechanism. B. thetaiotaomicron is reported to be able to signal to the host to produce more fucosylated glycoproteins (22). Although the mechanism remains unclear, it is argued that this signaling interaction is important in gaining preferential access to the substrate. Activation of a quorum-sensing system by fucose in R. inulinivorans may provide a crucial mechanism allowing successful competition for this host-derived substrate. Quorum-sensing mechanisms in gram-positive bacteria have thus far mainly been identified in pathogens associated with the expression of virulence genes and toxin production. However, a similar locus in Staphylococcus epidermis is involved in biofilm formation (42). This is the first identification of quorum-sensing genes in a commensal gastrointestinal anaerobe, and further work is required to identify the genes regulated by expression of this pathway. A two-component regulatory system was recently identified in the oral Lactobacillus plantarum strain WCFS1 (40) that also encodes a similar cyclic AIP. Among other functions this peptide regulates adherence of L. plantarum to a solid surface.
The main limitation of the shotgun microarray screening approach is the need for targeted sequencing to identify induced genes. Nevertheless, this work shows that the approach provides a powerful tool to investigate differential gene expression in unsequenced bacterial genomes, especially when comparing conditions that result in similar growth rates but that incorporate a single variable such as an alternative substrate. In this microarray screen most of the clones we analyzed were induced more than eightfold, and only a subset of those induced four- to eightfold were sequenced. Using this approach we were able to establish most of the gene products involved in fucose metabolism by R. inulinivorans A2-194 and determine that most of the relevant genes were tightly regulated. Importantly this analysis also revealed a potential mechanism by which these bacteria can communicate in response to changes in the surrounding environment, specifically the proximity of host-derived substrates.
Supplementary Material
Acknowledgments
We thank Pauline Young and Donna Henderson for DNA sequencing and David Brown for analysis of fermentation products. We also thank Sylvia Duncan, Perry Barrett, and Petra Louis for helpful discussions.
This work was funded by the Scottish Executive Environment and Rural Affairs Department (SEERAD).
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Badia, J., J. Ros, and J. Aguilar. 1985. Fermentation mechanism of fucose and rhamnose in Salmonella typhimurium and Klebsiella pneumoniae. J. Bacteriol. 161:435-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldoma, L., and J. Aguilar. 1988. Metabolism of l-fucose and l-rhamnose in Escherichia coli: aerobic-anaerobic regulation of l-lactaldehyde dissimilation. J. Bacteriol. 170:416-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barcenilla, A., S. E. Pryde, J. C. Martin, S. H. Duncan, C. S. Stewart, C. Henderson, and H. J. Flint. 2000. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl. Environ. Microbiol. 66:1654-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bobik, T. A., G. D. Havemann, R. J. Busch, D. S. Williams, and H. C. Aldrich. 1999. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B12-dependent 1,2-propanediol degradation. J. Bacteriol. 181:5967-5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan, W. C., B. J. Coyle, and P. Williams. 2004. Virulence regulation and quorum sensing in staphylococcal infections: competitive AgrC antagonists as quorum sensing inhibitors. J. Med. Chem. 47:4633-4641. [DOI] [PubMed] [Google Scholar]
- 6.Charrier, C., G. J. Duncan, M. D. Reid, G. J. Rucklidge, D. Henderson, P. Young, V. J. Russell, R. A. Aminov, H. J. Flint, and P. Louis. 2006. A novel class of CoA-transferase involved in short-chain fatty acid metabolism in butyrate-producing human colonic bacteria. Microbiology 152:179-185. [DOI] [PubMed] [Google Scholar]
- 7.Chen, P., D. I. Andersson, and J. R. Roth. 1994. The control region of the pdu/cob regulon in Salmonella typhimurium. J. Bacteriol. 176:5474-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coyne, M. J., B. Reinap, M. M. Lee, and L. E. Comstock. 2005. Human symbionts use a host-like pathway for surface fucosylation. Science 307:1778-1781. [DOI] [PubMed] [Google Scholar]
- 9.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:386-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duncan, S. H., R. I. Aminov, K. P. Scott, P. Louis, T. B. Stanton, and H. J. Flint. Proposal of three new species of Roseburia: Roseburia faecis (sp. nov.), Roseburia hominis (sp. nov.), Roseburia inulinivorans (sp. nov.), based on isolates from human faeces. Int. J. Syst. Evol. Microbiol., submitted for publication. [DOI] [PubMed]
- 11.Duncan, S. H., G. L. Hold, A. Barcenilla, C. S. Stewart, and H. J. Flint. 2002. Roseburia intestinalis sp. nov., a novel saccharolytic, butyrate-producing bacterium from human faeces. Int. J. Syst. Evol. Microbiol. 52:1615-1620. [DOI] [PubMed] [Google Scholar]
- 12.Duncan, S. H., K. P. Scott, A. G. Ramsay, H. J. Harmsen, G. W. Welling, C. S. Stewart, and H. J. Flint. 2003. Effects of alternative dietary substrates on competition between human colonic bacteria in an anaerobic fermentor system. Appl. Environ. Microbiol. 69:1136-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 308:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finne, J., M. E. Breimer, G. C. Hansson, K.-A. Karlsson, H. Leffler, J. F. G. Vliegenthart, and H. van Halbeek. 1989. Novel polyfucosylated N-linked glycopeptides with blood group A, H, X and Y determinants from small intestinal epithelial cells. J. Biol. Chem. 264:5720-5735. [PubMed] [Google Scholar]
- 15.Franks, A. H., H. J. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hacking, A. J., and E. C. Lin. 1977. Regulatory changes in the fucose system associated with the evolution of a catabolic pathway for propanediol in Escherichia coli. J. Bacteriol. 130:832-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartmanis, M. G., and T. C. Stadtman. 1986. Diol metabolism and diol dehydratase in Clostridium glycolicum. Arch. Biochem. Biophys. 245:144-152. [DOI] [PubMed] [Google Scholar]
- 18.Havemann, G. D., and T. A. Bobik. 2003. Protein content of polyhedral organelles involved in coenzyme B12-dependent degradation of 1,2-propanediol in Salmonella enterica serovar Typhimurium LT2. J. Bacteriol. 185:5086-5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayward, R. E., J. L. Derisi, S. Alfadhli, D. C. Kaslow, P. O. Brown, and P. K. Rathod. 2000. Shotgun DNA microarrays and stage-specific gene expression in Plasmodium falciparum malaria. Mol. Microbiol. 35:6-14. [DOI] [PubMed] [Google Scholar]
- 20.Hold, G. L., S. E. Pryde, V. J. Russell, E. Furrie, and H. J. Flint. 2002. Assessment of microbial diversity in human colonic samples by 16S rDNA sequence analysis. FEMS Microbiol. Ecol. 39:33-39. [DOI] [PubMed] [Google Scholar]
- 21.Hold, G. L., A. Schwiertz, R. I. Aminov, M. Blaut, and H. J. Flint. 2003. Oligonucleotide probes that detect quantitatively significant groups of butyrate-producing bacteria in human feces. Appl. Environ. Microbiol. 69:4320-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hooper, L. V., T. Midtvedt, and J. I. Gordon. 2002. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 22:283-307. [DOI] [PubMed] [Google Scholar]
- 23.Hooper, L. V., J. Xu, P. G. Falk, T. Midtvedt, and J. I. Gordon. 1999. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc. Natl. Acad. Sci. USA 96:9833-9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawrence, J. G., and J. R. Roth. 1996. Evolution of coenzyme B12 synthesis among enteric bacteria: evidence for loss and reacquisition of a multigene complex. Genetics 142:11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawrence, J. G., and J. R. Roth. 1996. Selfish operons: horizontal transfer may drive the evolution of gene clusters. Genetics 143:1843-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lay, C., M. Sutren, V. Rochet, K. Saunier, J. Dore, and L. Rigottier-Gois. 2005. Design and validation of 16S rRNA probes to enumerate members of the Clostridium leptum subgroup in human faecal microbiota. Environ. Microbiol. 7:933-946. [DOI] [PubMed] [Google Scholar]
- 27.Louis, P., S. H. Duncan, S. I. McCrae, J. Millar, M. S. Jackson, and H. J. Flint. 2004. Restricted distribution of the butyrate kinase pathway among butyrate-producing bacteria from the human colon. J. Bacteriol. 186:2099-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lyon, G. J., J. S. Wright, A. Christopoulos, R. P. Novick, and T. W. Muir. 2002. Reversible and specific extracellular antagonism of receptor-histidine kinase signalling. J. Biol. Chem. 277:6247-6253. [DOI] [PubMed] [Google Scholar]
- 29.Miyazaki, K., J. C. Martin, R. Marinsek-Logar, and H. J. Flint. 1997. Degradation and utilization of xylans by the rumen anaerobe Prevotella bryantii (formerly P. ruminicola subsp. brevis) B14. Anaerobe 3:373-381. [DOI] [PubMed] [Google Scholar]
- 30.Novick, R. P., S. J. Projan, J. Kornblum, H. F. Ross, G. Ji, B. Kreiswirth, F. Vandenesch, and S. Moghazeh. 1995. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol. Gen. Genet. 248:446-458. [DOI] [PubMed] [Google Scholar]
- 31.O'Brien, J. R., C. Raynaud, C. Croux, L. Girbal, P. Soucaille, and W. N. Lanzilotta. 2004. Insight into the mechanism of the B12-independent glycerol dehydratase from Clostridium butyricum: preliminary biochemical and structural characterization. Biochemistry 43:4635-4645. [DOI] [PubMed] [Google Scholar]
- 32.Ottenstein, D. M., and D. A. Bartley. 1971. Separation of free acids C2-C5 in dilute aqueous solution column technology. J. Chromatogr. Sci. 9:673-682. [Google Scholar]
- 33.Parro, V., and M. Moreno-Paz. 2003. Gene function analysis in environmental isolates: the nif regulon of the strict iron oxidizing bacterium Leptospirillum ferrooxidans. Proc. Natl. Acad. Sci. USA 100:7883-7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pitcher, M. C., and J. H. Cummings. 1996. Hydrogen sulphide: a bacterial toxin in ulcerative colitis? Gut 39:1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pryde, S. E., S. H. Duncan, G. L. Hold, C. S. Stewart, and H. J. Flint. 2002. The microbiology of butyrate formation in the human colon. FEMS Microbiol. Lett. 217:133-139. [DOI] [PubMed] [Google Scholar]
- 36.Ramsay, A. G. 2003. Studies on the molecular biology and ecology of butyrate-producing human colonic bacteria. Ph.D. thesis. University of Aberdeen, Aberdeen, UK.
- 37.Raynaud, C., P. Sarcabal, I. Meynial-Salles, C. Croux, and P. Soucaille. 2003. Molecular characterization of the 1,3-propanediol (1,3-PD) operon of Clostridium butyricum. Proc. Natl. Acad. Sci. USA 100:5010-5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salyers, A. A., J. R. Vercellotti, S. E. West, and T. D. Wilkins. 1977. Fermentation of mucin and plant polysaccharides by strains of Bacteroides from the human colon. Appl. Environ. Microbiol. 33:319-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Sturme, M. H., J. Nakayama, D. Molenaar, Y. Murakami, R. Kunugi, T. Fujii, E. E. Vaughan, M. Kleerebezem, and W. M. de Vos. 2005. An agr-like two-component regulatory system in Lactobacillus plantarum is involved in production of a novel cyclic peptide and regulation of adherence. J. Bacteriol. 187:5224-5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suau, A., R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Dore. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vuong, C., C. Gerke, G. A. Somerville, E. R. Fischer, and M. Otto. 2003. Quorum-sensing control of biofilm factors in Staphylococcus epidermidis. J. Infect. Dis. 188:706-718. [DOI] [PubMed] [Google Scholar]
- 43.Walker, A. W., S. H. Duncan, E. C. McWilliam Leitch, M. W. Child, and H. J. Flint. 2005. pH and peptide supply can radically alter bacterial populations and short-chain fatty acid ratios within microbial communities from the human colon. Appl. Environ. Microbiol. 71:3692-3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weisberg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu, J., M. K. Bjursell, J. Himrod, S. Deng, L. K. Carmichael, H. C. Chiang, L. V. Hooper, and J. I. Gordon. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074-2076. [DOI] [PubMed] [Google Scholar]
- 46.Yang, Y. H., S. Dudoit, P. Luu, D. M. Lin, V. Peng, J. Ngai, and T. P. Speed. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yarwood, J. M., and P. M. Schlievert. 2003. Quorum sensing in Staphylococcus infections. J. Clin. Investig. 112:1620-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, L., L. Gray, R. P. Novick, and G. Ji. 2002. Transmembrane topology of AgrB, the protein involved in the post-translational modification of AgrD in Staphylococcus aureus. J. Biol. Chem. 277:34736-34742. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, L., J. Lin, and G. Ji. 2004. Membrane anchoring of the AgrD N-terminal amphipathic region is required for its processing to produce a quorum-sensing pheromone in Staphylococcus aureus. J. Biol. Chem. 279:19448-19456. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.