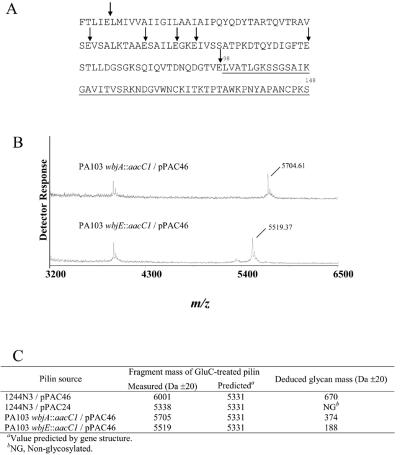
FIG. 5.
GluC digestion of 1244 pilin. (A) Amino acid sequence of 1244 pilin in which vertical arrows indicate GluC cleavage sites. The largest GluC fragment (underlined) consists of residues 98 to 148 and is predicted to have a molecular mass of 5,331 Da. This fragment contains the 1244 pilin glycosylation site, Ser148 (10). (B) MALDI-TOF spectra for GluC-treated pilin produced by PA103 wbjA::aacC1/pPAC46 and PA103 wbjE::aacC1/pPAC46 are shown. (C) The deduced glycan mass is listed for each of the pilins subjected to GluC digestion and MALDI-TOF.