Figure 2. Record Alignment and Pause Locations Identified.
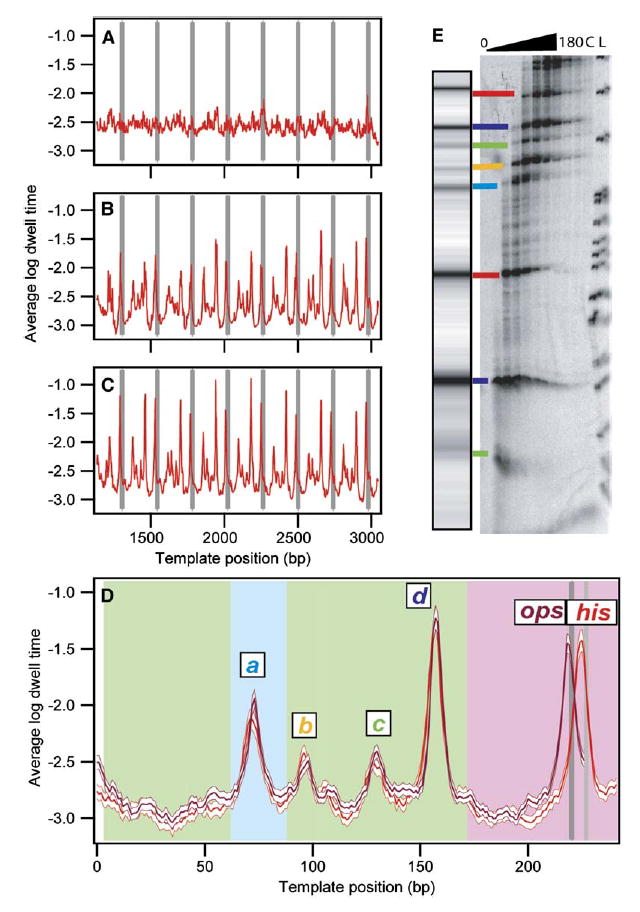
(A–C) Dwell-time histograms were compiled for each transcriptional record as a function of position. The logarithms of these histograms were then averaged for groups of records, as follows: (A) Average log dwell-time histogram for his records (n = 53) before any rescaling or offsets applied.
(B) Average log dwell-time histogram for terminating his traces (n = 27) after initial rescaling and alignment of records at the termination site. (C) Average log dwell-time histogram for all his traces (n = 53) after final alignment.
(D) Average log dwell-time histogram for aligned data computed from all eight repeats for the his repeat motif (red; n = 53 molecules, 310 records) and the ops repeat motif (magenta; n = 61 molecules, 419 records), shown with the bootstrapped standard deviations (white). Background color indicates origin of the underlying sequences: rpoB gene (green), restriction sites used for cloning (light blue), regulatory pause region (pink), ops pause site (dark gray), his pause site (light gray). Major pause sites are labeled.
(E) Comparison of single-molecule and bulk transcription data. A simulated transcription gel was created from the dataset in (D) using a grayscale proportional to the peak height and scaling the position logarithmically to approximate RNA gel mobility. [α-32P]GMP-labeled transcription complexes were incubated with 250 μM NTPs, quenched at times between 0 s and 180 s, and run on a denaturing polyacrylamide gel. Lane L shows the MspI pBR322 ladder; lane C is a chase. Lines are drawn between corresponding bands identified in single-molecule and gel data, color coded as in (D).
