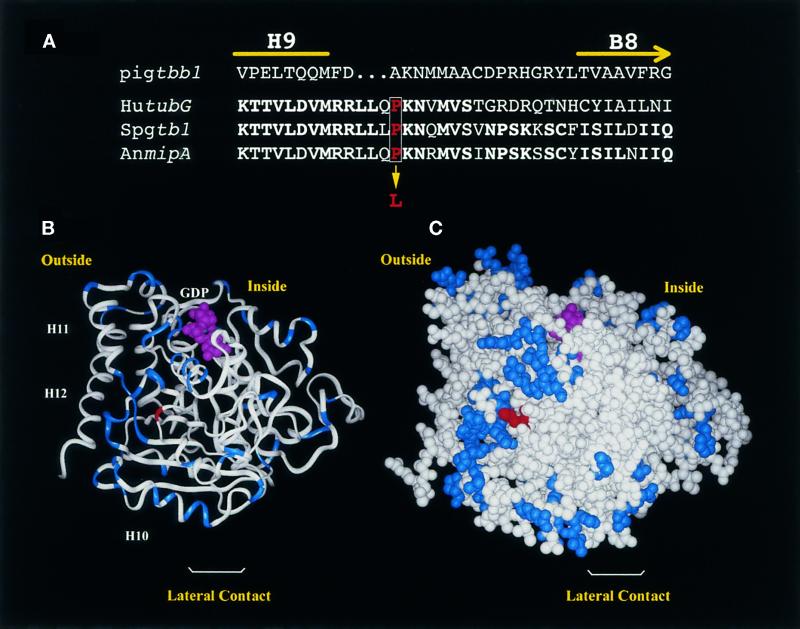
Figure 3.
Predicted structure of γ-tubulin showing the position of the SL1 mutation. (A) Partial sequence alignment of pig β-tubulin (Pigtbb1) and γ-tubulin from human (hutubG), S. pombe (Spgtb1), and A. nidulans (AnmipA) showing amino acids from H9 through B8. This corresponds to amino acids 298–324 in Spgtb1. Identical amino acids are in bold. The proline (red type, boxed) that is mutated in SL1 to a leucine is indicated. (B and C) Predicted 3D structure of γ-tubulin, obtained by replacing differing amino acids in pig α-tubulin with the corresponding γ-tubulin amino acids. Residues that are similar in α- or β-tubulin but that have a totally different character in γ-tubulin (e.g., small vs. large or hydrophobic vs. charged) are highlighted in blue. The PL301 mutation is shown in red. (B) Ribbon diagram. The nucleotide and the last C-terminal helices (H10, H11, and H12) are indicated. Klps have been reported to bind at H11 and H12 of β-tubulin. (C) CPK representation. The orientation is such that, if attached to a microtubule end, the viewer will be facing a site of lateral interaction. The inside and outside of the microtubule are indicated.