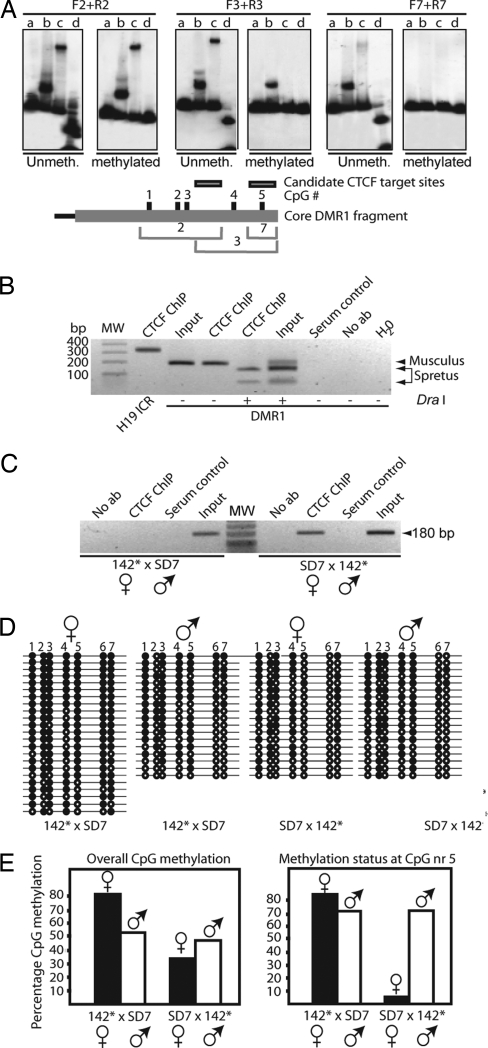
Fig. 4.
ICR–DMR1, CTCF, and long-range epigenetic coordination. (A) (Upper) Effects of in vitro CpG methylation on CTCF binding to positive DNA fragments 2, 3, and 7 (the last fragment covers only CpG site nr 5; see Fig. 13). Digestion of unmethylated/methylated probe with the methylation-sensitive enzyme HpaII is depicted in lane d for each panel. Three pair of panels with both unmethylated and methylated [32P]DNA probes and in vitro-translated proteins are depicted (lanes are marked as in Fig. 13 Upper). (Lower) Shown is a map of core DMR1 fragment with CpG numbers and the positions of the overlapping DNA fragments. (B and C) CTCF interacts with the Igf2 DMR1 in vivo. ChIP analysis of neonatal liver derived from a cross between C57BL/6 and SD7 (B) or from 142* × SD7 and SD7 × 142* crosses (C). The DraI polymorphism specific for the M. spretus allele of the DMR1 in the recombinant SD7 mouse strain was exploited (see Fig. 3A). (D) Bisulfite sequencing data of DMR1 in neonatal liver. Filled and open circles represent methylated and unmethylated CpGs, respectively. Maternal and paternal alleles were distinguished using the DraI polymorphism. (E) Summary of the overall bisulfite data or, specifically, the fifth CpG site, given in percentage of CpG methylation.