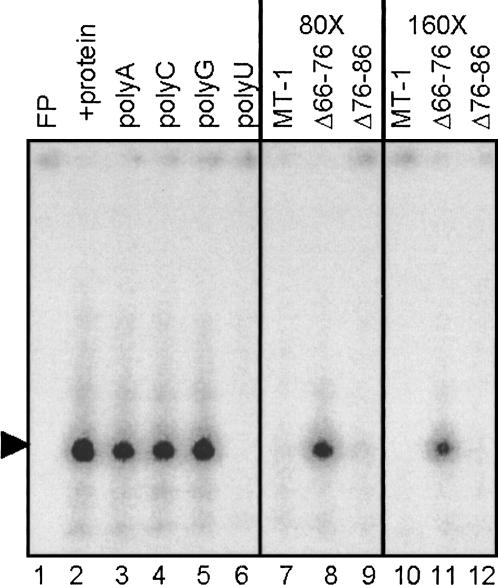
FIGURE 2.
RNA–protein complex formation monitored by gel retardation assay. Complex formation was studied using [α-32P]CTP-labeled MT-1 3′UTR RNA (12 fmol) and 2 μg S100 extract protein from CHO cells and prepared as described in Materials and Methods. RNase T1 digestion was performed after the binding reaction and products separated by native PAGE. Lane 1 contains MT-1 3′UTR RNA alone (free probe; FP) and lane 2 shows formation of retardation complexes in the presence of cell extract. The effects of competition with a 100-fold mass excess of homoribopolymers polyA, C, G, and U are shown in lanes 3–6, respectively. Poly U dramatically reduces the major gel retardation complex. In lanes 7–12, either an 80- or 160-fold molar excess of MT-1, Δ66–76, and Δ76–86 unlabeled RNAs was used to compete with MT-1 RNA for protein binding. Note that MT-1 and Δ76–86 compete more effectively than Δ66–76.