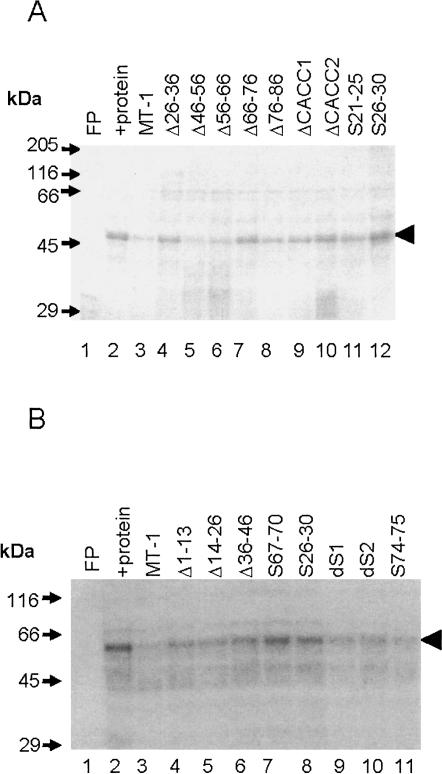
FIGURE 6.
UV cross-linking analysis of the effects of deletions and base substitutions on protein binding to MT-1 3′UTR. An [α-32P] CTP-labeled MT-1 3′UTR RNA probe (50 fmol) was incubated with 35 μg protein (CHO cell S100 extract) prepared as described in Materials and Methods and 160× molar excess of unlabeled competitor transcripts. Complexes were fixed by UV irradiation, and the probe was removed by RNase A digestion. Labeled proteins were separated by SDS-PAGE and detected by autoradiography (A,B). In the absence of protein there was no binding detected (free probe; FP). Note the major labeled band of ∼50 kDa (arrowhead) after incubation of the probe with protein extract (+protein). Binding to this protein is effectively competed by a 160× molar excess of unlabeled competitor MT-1 3′UTR (A, B, lane 3) but markedly less so by Δ66–76, Δ26–36, ΔCACC1, ΔCACC2, S21–25, S26–30 (A, lanes 4,7,9–12), S67–70, and S26–30 (B, lanes 7,8) (A, B; transcript abbreviations as shown in Fig. 1). Transcripts Δ46–56, Δ56–66, Δ76–86 (A, lanes 5,6,8) dS1, dS2, and S74–75 (B, lanes 9–11) competed effectively for protein binding.