TABLE 2.
Yeast HisRS aminoacylation of microhelixHis variants
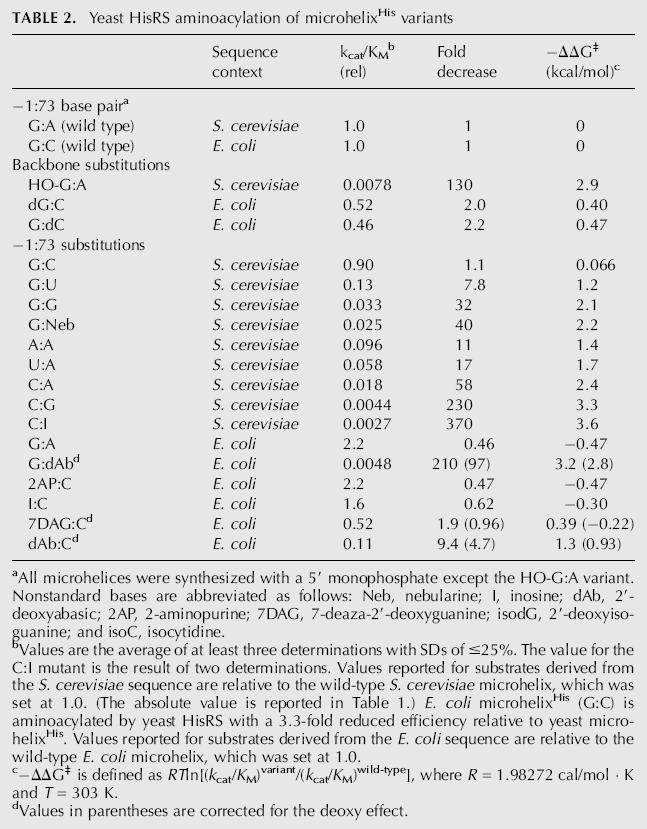
aAll microhelices were synthesized with a 5′ monophosphate except the HO-G:A variant. Nonstandard bases are abbreviated as follows: Neb, nebularine; I, inosine; dAb, 2′-deoxyabasic; 2AP, 2-aminopurine; 7DAG, 7-deaza-2′-deoxyguanine; isodG, 2′-deoxyisoguanine; and isoC, isocytidine.
bValues are the average of at least three determinations with SDs of ≤25%. The value for the C:I mutant is the result of two determinations. Values reported for substrates derived from the S. cerevisiae sequence are relative to the wild-type S. cerevisiae microhelix, which was set at 1.0. (The absolute value is reported in Table 1.) E. coli microhelixHis (G:C) is aminoacylated by yeast HisRS with a 3.3-fold reduced efficiency relative to yeast microhelixHis. Values reported for substrates derived from the E. coli sequence are relative to the wild-type E. coli microhelix, which was set at 1.0.
c−ΔΔG‡ is defined as RTln[(k cat/K M)variant/(k cat/K M)wild-type], where R = 1.98272 cal/mol ;· ;K and T = 303 K.
dValues in parentheses are corrected for the deoxy effect.
