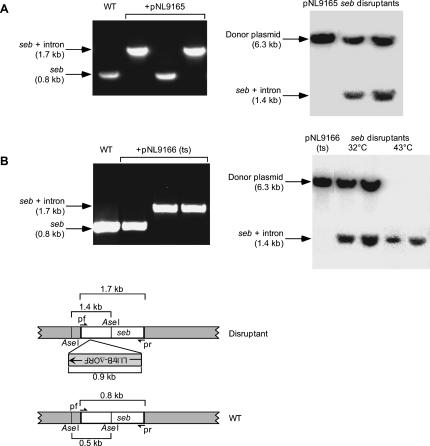
FIGURE 4.
Use of the Ll.LtrB targetron to disrupt the S. aureus seb gene. (A,B) Disruption of the seb gene in S. aureus RN8098 using targetron Seb-123a expressed from pNL9165 and pNL9166. For all steps, cells containing pNL9165 (pCN37-derivative) were grown at 37°C, and those containing pNL9166 (pCN39-derivative; temperature-sensitive replicon) were grown at 32°C. Donor plasmids were electroporated into S. aureus RN4220, then reisolated and electroporated into RN8098 (see Materials and Methods). Cells were grown in BHI medium containing erythromycin, then induced with 10 mM CdCl2 for 90 min, and plated on BHI medium containing erythromycin. The left panels show colony PCR of wild-type RN8098 (WT) and representative colonies obtained with each of the donor plasmids. The right panels show Southern blots of AseI-digested donor plasmids pNL9165 and pNL9166 (left lane) and randomly chosen seb disruptants obtained with the corresponding donor plasmid (right lane). The blots were hybridized with a 32P-labeled intron probe. The samples in the two rightmost lanes of the Southern blot in panel B are from cells in which pNL9166 was cured from the disruptants by overnight incubation at 43°C on BHI plates. The bands shown are the only ones visible on the blots (molecular weight range 100 bp–12 kb). The schematic at the bottom shows a diagram of the seb gene, with (top) and without (bottom) the integrated targetron, including the location of primers pf and pr used to detect targetron integration by colony PCR and the location of the AseI sites used for Southern hybridization analysis.