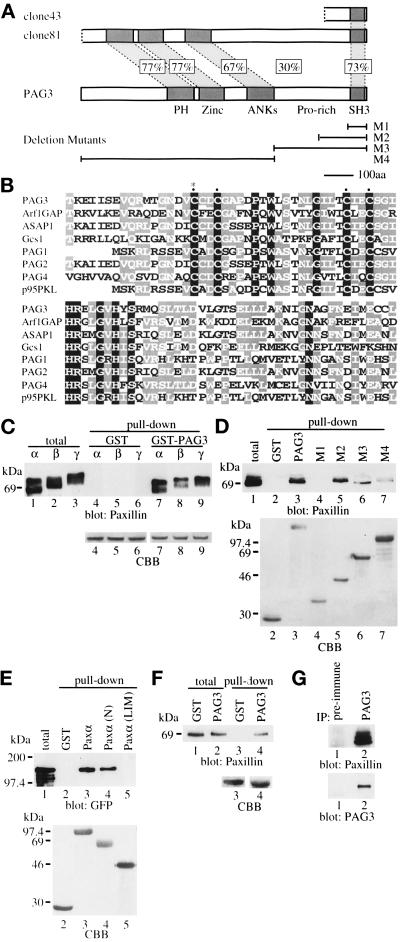
Figure 1.
Structure of PAG3 and its binding toward paxillin. (A) Schematic diagram of PAG3 and its comparison with original two clones isolated using GST-paxillin α as a probe (clones 43 and 81). (B) Comparison of the zinc finger domain of PAG3 (residues 420–505) with those of ARF1 GAP (residues 6–91), ASAP1 (residues 453–538; Brown et al., 1998), GCS1 (residues 10–95), PAG1 (residues 1–83), PAG2 (which corresponds to clone 81), PAG4 (residues 404– 489), and p95PKL (residues 1–83). The cDNA for PAG2 was not complete; therefore, residue numbers were not assigned. Identical residues are framed and shadowed. The positions of four conserved cysteines of the zinc finger motif are marked by dots, and the residue mutated in the PAG3 CA mutant is marked by an asterisk. ASAP1 corresponds to the s19 clone, and PAG1, PAG2, and PAG4 correspond to our collection of paxillin-binding proteins (our unpublished data). (C) PAG3 binding to paxillin isoforms. Each 2.5 μg of GST alone (lanes 4–6) or a GST fusion form of PAG3 (lanes 7–9) expressed in COS-7 cells and purified on glutathione beads were incubated with 15 μg of Sf-9 cell lysates, each producing recombinant paxillin isoforms of α, β, and γ. After incubation, beads were washed, and proteins were retained on the beads were subjected to immunoblotting analysis using anti-paxillin antibody, Ab 199–217. Each 0.5 μg of the same cell lysates was included as “total” (lanes 1–3). (D) Paxillin binds to the COOH-terminal region of PAG3. Each 2.5 μg of GST alone (lane 2) or GST fusion forms of wild-type and deletion mutants of PAG3 (lanes 3–7) purified on glutathione beads as described in MATERIALS AND METHODS were incubated with 15 μg of Sf-9 cell lysate producing recombinant paxillin α to test binding, as above. Lane 1 included 0.5 μg of the same cell lysate. For the deletion mutants of M1–M4, see A. (E) PAG3 binds to the NH2-terminal region of paxillin. Each 5 μg of GST alone (lane 2) and GST fusion forms of paxillin wild type and mutants (lanes 3–5) as described in MATERIALS AND METHODS were purified on glutathione beads and incubated with 500 μg of COS-7 cell lysate expressing the EGFP fusion form of PAG3 to test binding. Thirty micrograms of the total COS-7 cell lysate were included in lane 1. Immunoblot was done with anti-GFP antibody. (F) Association of PAG3 and paxillin in vivo. Each 1 × 106 COS-7 cells were transfected with 10 μg of pEBG or pEBG/PAG3 plasmid, and GST (lane 3) or GST-PAG3 (lane 4) was pulled down from each 1 mg of the cell lysate using glutathione beads to analyze its association with endogenous paxillin. Each 30 μg of the total cell lysates were included in lane 1 (cells with pEBG) and lane 2 (cells with pEBG/PAG3). Immunoblot was done with anti-paxillin antibody, Ab 199–217. (G) In vivo association of endogenous PAG3 and paxillin. Each 1 mg of cell lysate prepared from TPA-treated U937 cells was subjected to immunoprecipitation using anti-PAG3 antibody (lane 2) or the preimmune serum (lane 1) coupled with protein A-Sepharose beads. Precipitated proteins were then subjected to immunoblotting analysis using anti-paxillin antibody, Ab 199–217, and anti-PAG3 antibody. In C–F, amounts of each fusion protein used for pull-down assays are shown by Coomassie brilliant blue staining (CBB).