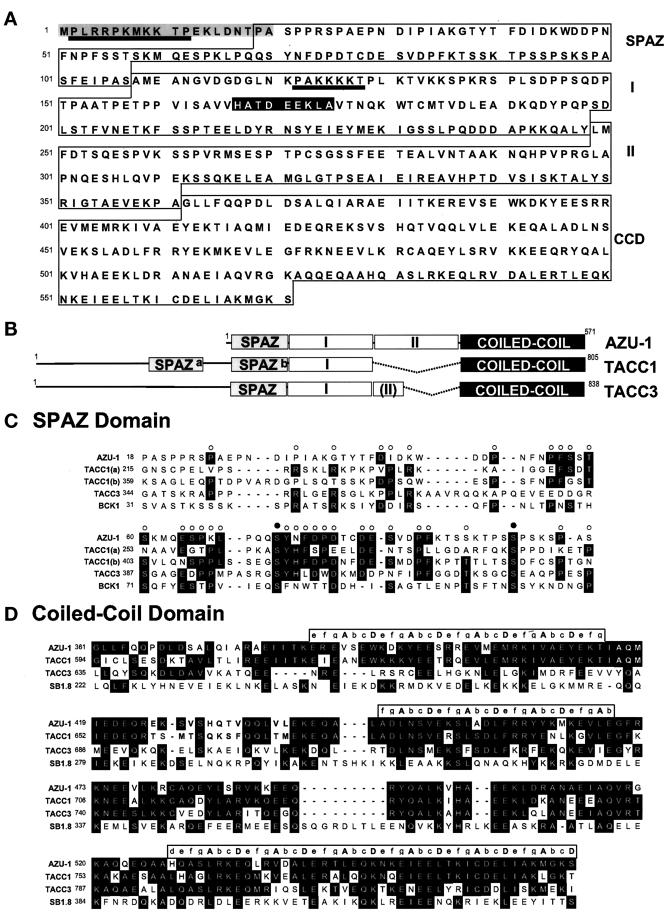
Figure 2.
Sequence and structure of AZU-1. (A) Deduced amino acid sequence of the AZU-1 571-amino-acid open reading frame. Four structural domains, labeled SPAZ, region I, region II, and CCD, are boxed, and two predicted NLS motifs are underlined. The N-terminal peptide used to generate the AZU-1 antibody is highlighted in gray. The HATDEEKLA sequence, a peptide conserved between AZU-1 and TACC1, appears in black. (B) Domain organization of AZU-1 and two AZU-1-related genes, TACC1 and TACC3. Based on its similarity with TACC1 and TACC3, AZU-1 can be partitioned into four domains: 1) the N-terminal SPAZ domain, 2) region I, a region that shares a moderate sequence similarity with TACC1 and to a lesser extent with TACC3, 3) region II, which is totally absent in TACC1 and partially removed from TACC3, and 4) the C-terminal coiled-coil domain. (C) Sequence alignments of SPAZ domains from AZU-1, TACC1 (2 copies, a and b), TACC3, and BCK1 from S. cerevisiae. Residues that are conserved in three or more of these sequences appear in black; the corresponding columns are marked with open circles. Two invariant serine residues are indicated by filled circles. Fold recognition analyses predict that SPAZ domains adopt Ig-like folds. (D) CCD sequence alignments of AZU-1, TACC1, TACC3, and SB1.8/DXS423E. Amino acid identities observed in two or more of the aligned sequences are indicated in black; in cases in which two pairs of identical amino acids are observed in the alignment, AZU-1-like sequences are preferentially highlighted. The CCD heptad repeat positions, a–g, are indicated in brackets above the three regions where all four proteins fall into register. Positions a and d, often occupied by hydrophobic residues, are indicated in capital letters. Sequence identities among all four proteins in this region are most notable in the second half of the CCD.